Abstract
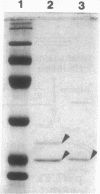
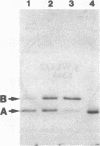
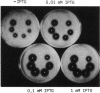
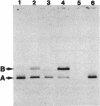
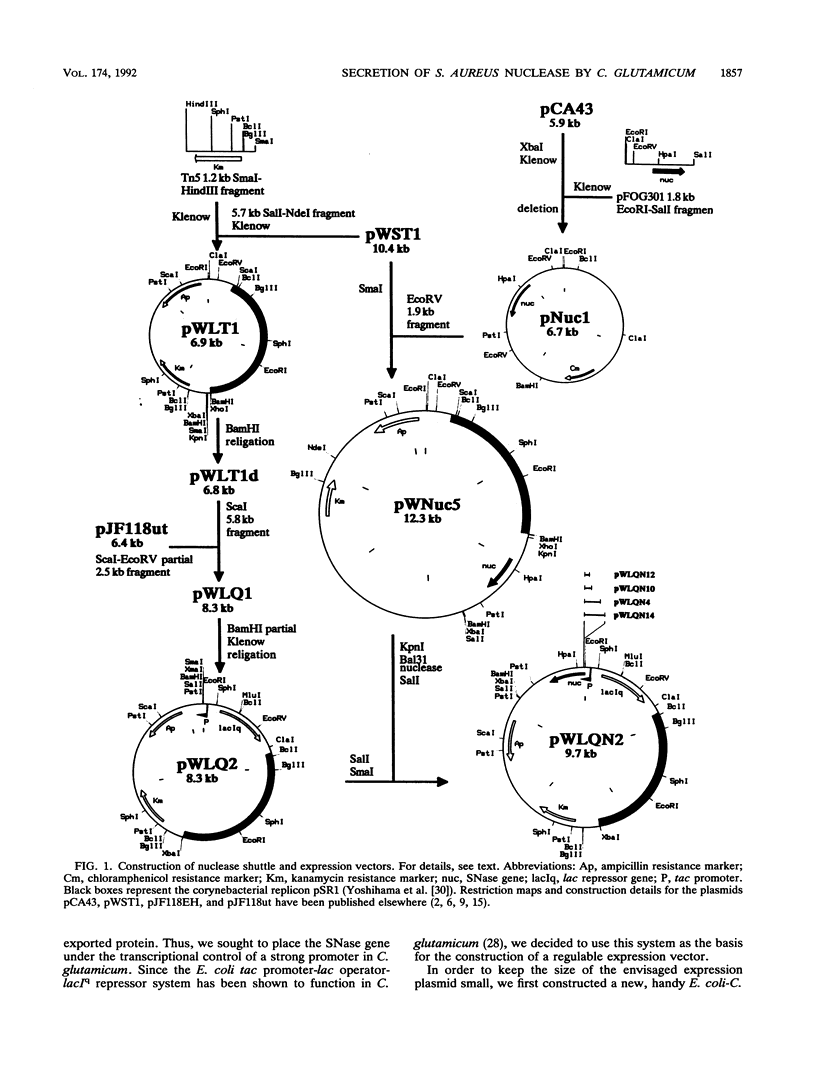
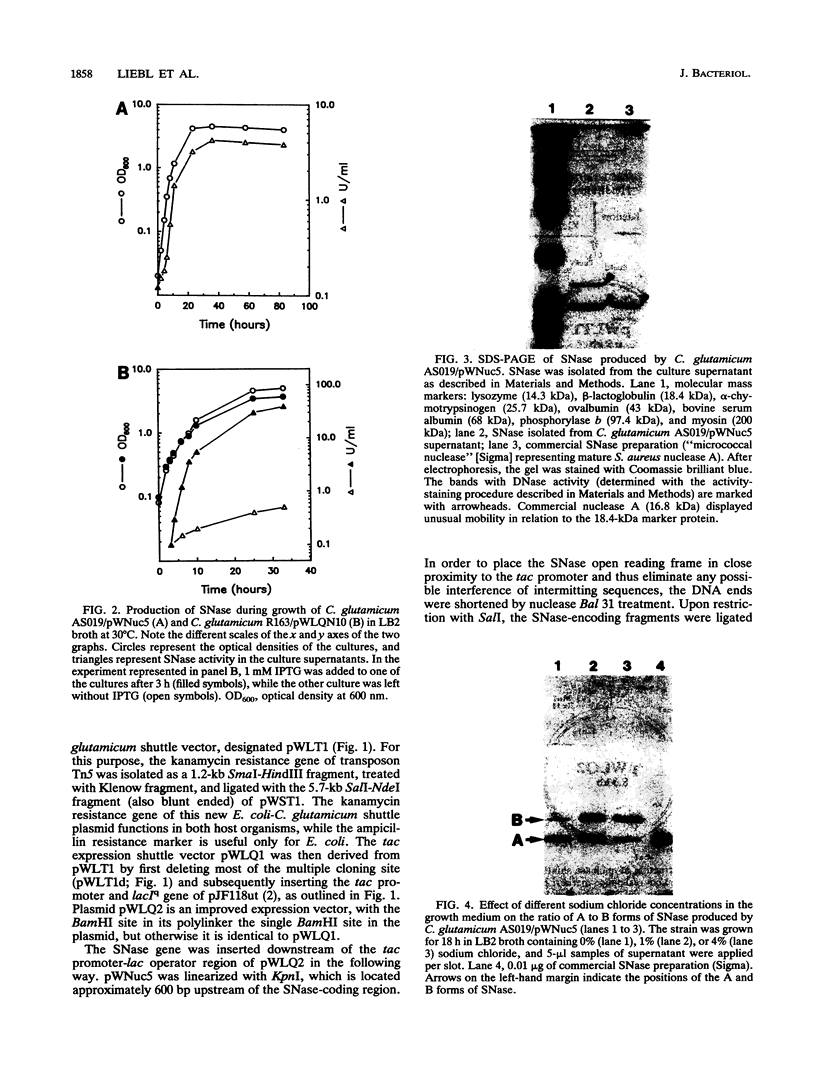
The gene for staphylococcal nuclease (SNase), an extracellular enzyme of Staphylococcus aureus, was introduced into Corynebacterium glutamicum. The heterologous gene was expressed in this host organism, and SNase was efficiently exported to the culture medium. Amino-terminal sequencing of SNase secreted by C. glutamicum revealed that the signal peptide was apparently cleaved off at precisely the same position as in the original host, S. aureus. As with S. aureus, a second smaller form of SNase (A form), whose appearance is presumably the result of a secondary processing step, was found in the culture medium of the recombinant C. glutamicum strain. The A form was one residue shorter than the mature nuclease A produced by S. aureus. Variation of the sodium chloride concentration in the growth medium had a marked influence on the location and the processing of SNase by C. glutamicum. In a complex growth medium containing 4% sodium chloride, SNase was exclusively located in the supernatant, but a significant amount of the enzyme remained cell associated if the strain was grown in a low-salt medium. Also, high salt concentrations seemed to inhibit processing of the high-molecular-weight form of SNase (B form) to the smaller A form. Similarities and differences in the export and modes of processing of SNase by three different, nonrelated gram-positive host organisms are discussed. Finally, a versatile Escherichia coli-C. glutamicum tac-lacIq expression shuttle vector was constructed. With this vector, it was possible to achieve isopropyl-beta-D-galactopyranoside (IPTG)-inducible overexpression and secretion of SNase in C. glutamicum, whereby the expression level was dependent on the concentration of the inducer.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cone J. L., Cusumano C. L., Taniuchi H., Anfinsen C. B. Staphylococcal nuclease (Foggi strain). II. The amino acid sequence. J Biol Chem. 1971 May 25;246(10):3103–3110. [PubMed] [Google Scholar]
- Cuatrecasas P., Fuchs S., Anfinsen C. B. Catalytic properties and specificity of the extracellular nuclease of Staphylococcus aureus. J Biol Chem. 1967 Apr 10;242(7):1541–1547. [PubMed] [Google Scholar]
- Davis A., Moore I. B., Parker D. S., Taniuchi H. Nuclease B. A possible precursor of nuclease A, an extracellular nuclease of Staphylococcus aureus. J Biol Chem. 1977 Sep 25;252(18):6544–6553. [PubMed] [Google Scholar]
- Fürste J. P., Pansegrau W., Frank R., Blöcker H., Scholz P., Bagdasarian M., Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48(1):119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- Gierasch L. M. Signal sequences. Biochemistry. 1989 Feb 7;28(3):923–930. doi: 10.1021/bi00429a001. [DOI] [PubMed] [Google Scholar]
- Kovacevic S., Veal L. E., Hsiung H. M., Miller J. R. Secretion of staphylococcal nuclease by Bacillus subtilis. J Bacteriol. 1985 May;162(2):521–528. doi: 10.1128/jb.162.2.521-528.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreutz B., Götz F. Construction of Staphylococcus plasmid vector pCA43 conferring resistance to chloramphenicol, arsenate, arsenite and antimony. Gene. 1984 Nov;31(1-3):301–304. doi: 10.1016/0378-1119(84)90226-9. [DOI] [PubMed] [Google Scholar]
- Lachica R. V., Genigeorgis C., Hoeprich P. D. Metachromatic agar-diffusion methods for detecting staphylococcal nuclease activity. Appl Microbiol. 1971 Apr;21(4):585–587. doi: 10.1128/am.21.4.585-587.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebl W., Bayerl A., Schein B., Stillner U., Schleifer K. H. High efficiency electroporation of intact Corynebacterium glutamicum cells. FEMS Microbiol Lett. 1989 Dec;53(3):299–303. doi: 10.1016/0378-1097(89)90234-6. [DOI] [PubMed] [Google Scholar]
- Liebl W., Ehrmann M., Ludwig W., Schleifer K. H. Transfer of Brevibacterium divaricatum DSM 20297T, "Brevibacterium flavum" DSM 20411, "Brevibacterium lactofermentum" DSM 20412 and DSM 1412, and Corynebacterium glutamicum and their distinction by rRNA gene restriction patterns. Int J Syst Bacteriol. 1991 Apr;41(2):255–260. doi: 10.1099/00207713-41-2-255. [DOI] [PubMed] [Google Scholar]
- Liebl W., Götz F. Studies on lipase directed export of Escherichia coli beta-lactamase in Staphylococcus carnosus. Mol Gen Genet. 1986 Jul;204(1):166–173. doi: 10.1007/BF00330205. [DOI] [PubMed] [Google Scholar]
- Liss L. R., Johnson B. L., Oliver D. B. Export defect adjacent to the processing site of staphylococcal nuclease is suppressed by a prlA mutation. J Bacteriol. 1985 Nov;164(2):925–928. doi: 10.1128/jb.164.2.925-928.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987 Jul 25;262(21):10035–10038. [PubMed] [Google Scholar]
- Miller J. R., Kovacevic S., Veal L. E. Secretion and processing of staphylococcal nuclease by Bacillus subtilis. J Bacteriol. 1987 Aug;169(8):3508–3514. doi: 10.1128/jb.169.8.3508-3514.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabayaski K., Mizuno D. Surface-bound nuclease of Staphylococcus aureus: localization of the enzyme. J Bacteriol. 1974 Jan;117(1):215–221. doi: 10.1128/jb.117.1.215-221.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortle D. A genetic system for analysis of staphylococcal nuclease. Gene. 1983 May-Jun;22(2-3):181–189. doi: 10.1016/0378-1119(83)90102-6. [DOI] [PubMed] [Google Scholar]
- Smith M. D., Flickinger J. L., Lineberger D. W., Schmidt B. Protoplast transformation in coryneform bacteria and introduction of an alpha-amylase gene from Bacillus amyloliquefaciens into Brevibacterium lactofermentum. Appl Environ Microbiol. 1986 Mar;51(3):634–639. doi: 10.1128/aem.51.3.634-639.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahara M., Hibler D. W., Barr P. J., Gerlt J. A., Inouye M. The ompA signal peptide directed secretion of Staphylococcal nuclease A by Escherichia coli. J Biol Chem. 1985 Mar 10;260(5):2670–2674. [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Yoshihama M., Higashiro K., Rao E. A., Akedo M., Shanabruch W. G., Follettie M. T., Walker G. C., Sinskey A. J. Cloning vector system for Corynebacterium glutamicum. J Bacteriol. 1985 May;162(2):591–597. doi: 10.1128/jb.162.2.591-597.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]