Abstract
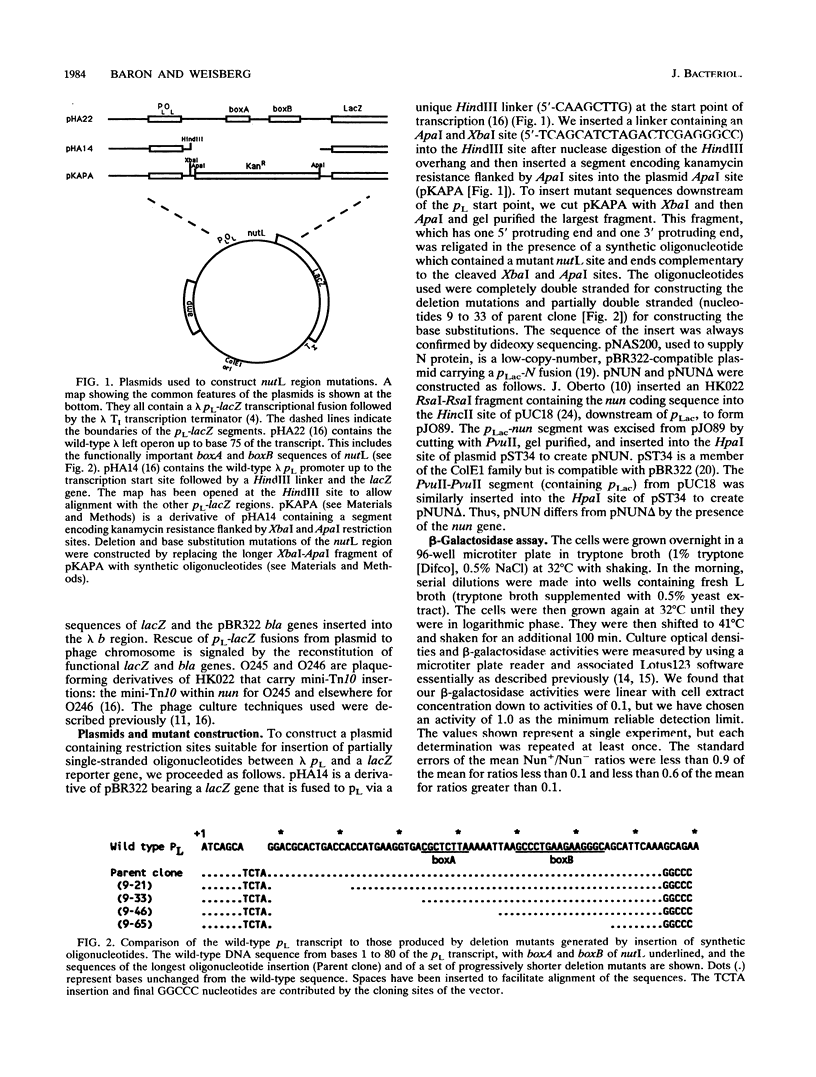
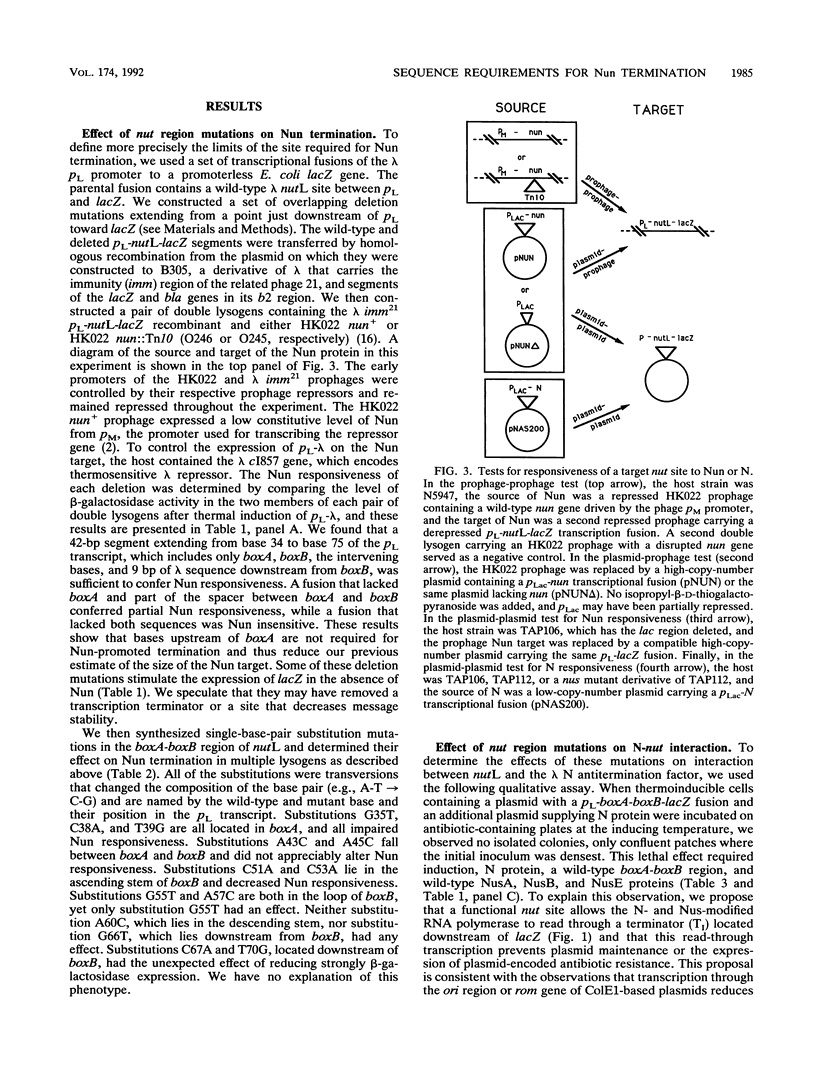
Phage HK022 encodes a protein, Nun, that promotes transcription termination within the pL and pR operons of its relative, phage lambda. The lambda sequences required for termination had previously been shown to overlap the nut sites, which are essential for transcription antitermination during normal lambda growth. To further specify the Nun target and to determine its relation to the nut sites, we constructed deletion and base substitution mutations of the lambda nutL region and measured Nun-dependent reduction of the expression of a downstream reporter gene. The shortest construct that retained full Nun responsiveness was a 42-bp segment that included both boxA and boxB, sequences that have been implicated in lambda antitermination. Deletion of boxA reduced Nun termination, and deletion of both sequences eliminated Nun termination. Base substitutions in boxA and the proximal portion of boxB impaired Nun termination, while base substitutions between boxA and boxB, in the distal portion of boxB, and immediately downstream from boxB had no appreciable effect. The termination defect of all of the base substitution mutations was relieved by increasing the level of Nun protein; in contrast, the deletions and a multiple-base substitution did not regain full Nun responsiveness at elevated Nun concentrations. We also asked if these mutant nut regions retained their ability to interact with N, the lambda-encoded antitermination protein. A qualitative assay showed that mutations within boxA or boxB reduced interaction, while mutations outside boxA and boxB did not. These data show that (i) the recognition sites for N and Nun overlap to a very considerable extent but are probably not identical and (ii) a high concentration of Nun promotes its interaction with mutant nut sites, a behavior also reported to be characteristic of N.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cam K. M., Oberto J., Weisberg R. A. The early promoters of bacteriophage HK022: contrasts and similarities to other lambdoid phages. J Bacteriol. 1991 Jan;173(2):734–740. doi: 10.1128/jb.173.2.734-740.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doelling J. H., Franklin N. C. Effects of all single base substitutions in the loop of boxB on antitermination of transcription by bacteriophage lambda's N protein. Nucleic Acids Res. 1989 Jul 25;17(14):5565–5577. doi: 10.1093/nar/17.14.5565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin N. C., Doelling J. H. Overexpression of N antitermination proteins of bacteriophages lambda, 21, and P22: loss of N protein specificity. J Bacteriol. 1989 May;171(5):2513–2522. doi: 10.1128/jb.171.5.2513-2522.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman D. I., Olson E. R., Johnson L. L., Alessi D., Craven M. G. Transcription-dependent competition for a host factor: the function and optimal sequence of the phage lambda boxA transcription antitermination signal. Genes Dev. 1990 Dec;4(12A):2210–2222. doi: 10.1101/gad.4.12a.2210. [DOI] [PubMed] [Google Scholar]
- Horwitz R. J., Li J., Greenblatt J. An elongation control particle containing the N gene transcriptional antitermination protein of bacteriophage lambda. Cell. 1987 Nov 20;51(4):631–641. doi: 10.1016/0092-8674(87)90132-2. [DOI] [PubMed] [Google Scholar]
- Lazinski D., Grzadzielska E., Das A. Sequence-specific recognition of RNA hairpins by bacteriophage antiterminators requires a conserved arginine-rich motif. Cell. 1989 Oct 6;59(1):207–218. doi: 10.1016/0092-8674(89)90882-9. [DOI] [PubMed] [Google Scholar]
- Menzel R. A microtiter plate-based system for the semiautomated growth and assay of bacterial cells for beta-galactosidase activity. Anal Biochem. 1989 Aug 15;181(1):40–50. doi: 10.1016/0003-2697(89)90391-6. [DOI] [PubMed] [Google Scholar]
- Oberto J., Weisberg R. A., Gottesman M. E. Structure and function of the nun gene and the immunity region of the lambdoid phage HK022. J Mol Biol. 1989 Jun 20;207(4):675–693. doi: 10.1016/0022-2836(89)90237-4. [DOI] [PubMed] [Google Scholar]
- Olson E. R., Tomich C. S., Friedman D. I. The nusA recognition site. Alteration in its sequence or position relative to upstream translation interferes with the action of the N antitermination function of phage lambda. J Mol Biol. 1984 Dec 25;180(4):1053–1063. doi: 10.1016/0022-2836(84)90270-5. [DOI] [PubMed] [Google Scholar]
- Oppenheim A. B., Gottesman S., Gottesman M. Regulation of bacteriophage lambda int gene expression. J Mol Biol. 1982 Jul 5;158(3):327–346. doi: 10.1016/0022-2836(82)90201-7. [DOI] [PubMed] [Google Scholar]
- Robert J., Sloan S. B., Weisberg R. A., Gottesman M. E., Robledo R., Harbrecht D. The remarkable specificity of a new transcription termination factor suggests that the mechanisms of termination and antitermination are similar. Cell. 1987 Nov 6;51(3):483–492. doi: 10.1016/0092-8674(87)90644-1. [DOI] [PubMed] [Google Scholar]
- Robledo R., Atkinson B. L., Gottesman M. E. Escherichia coli mutations that block transcription termination by phage HK022 Nun protein. J Mol Biol. 1991 Aug 5;220(3):613–619. doi: 10.1016/0022-2836(91)90104-e. [DOI] [PubMed] [Google Scholar]
- Robledo R., Gottesman M. E., Weisberg R. A. Lambda nutR mutations convert HK022 Nun protein from a transcription termination factor to a suppressor of termination. J Mol Biol. 1990 Apr 20;212(4):635–643. doi: 10.1016/0022-2836(90)90226-c. [DOI] [PubMed] [Google Scholar]
- Schauer A. T., Carver D. L., Bigelow B., Baron L. S., Friedman D. I. lambda N antitermination system: functional analysis of phage interactions with the host NusA protein. J Mol Biol. 1987 Apr 20;194(4):679–690. doi: 10.1016/0022-2836(87)90245-2. [DOI] [PubMed] [Google Scholar]
- Som T., Tomizawa J. Origin of replication of Escherichia coli plasmid RSF 1030. Mol Gen Genet. 1982;187(3):375–383. doi: 10.1007/BF00332615. [DOI] [PubMed] [Google Scholar]
- Stueber D., Bujard H. Transcription from efficient promoters can interfere with plasmid replication and diminish expression of plasmid specified genes. EMBO J. 1982;1(11):1399–1404. doi: 10.1002/j.1460-2075.1982.tb01329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen W. A., Das A. Action of an RNA site at a distance: role of the nut genetic signal in transcription antitermination by phage-lambda N gene product. New Biol. 1990 Nov;2(11):975–991. [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]


