Abstract
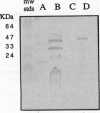
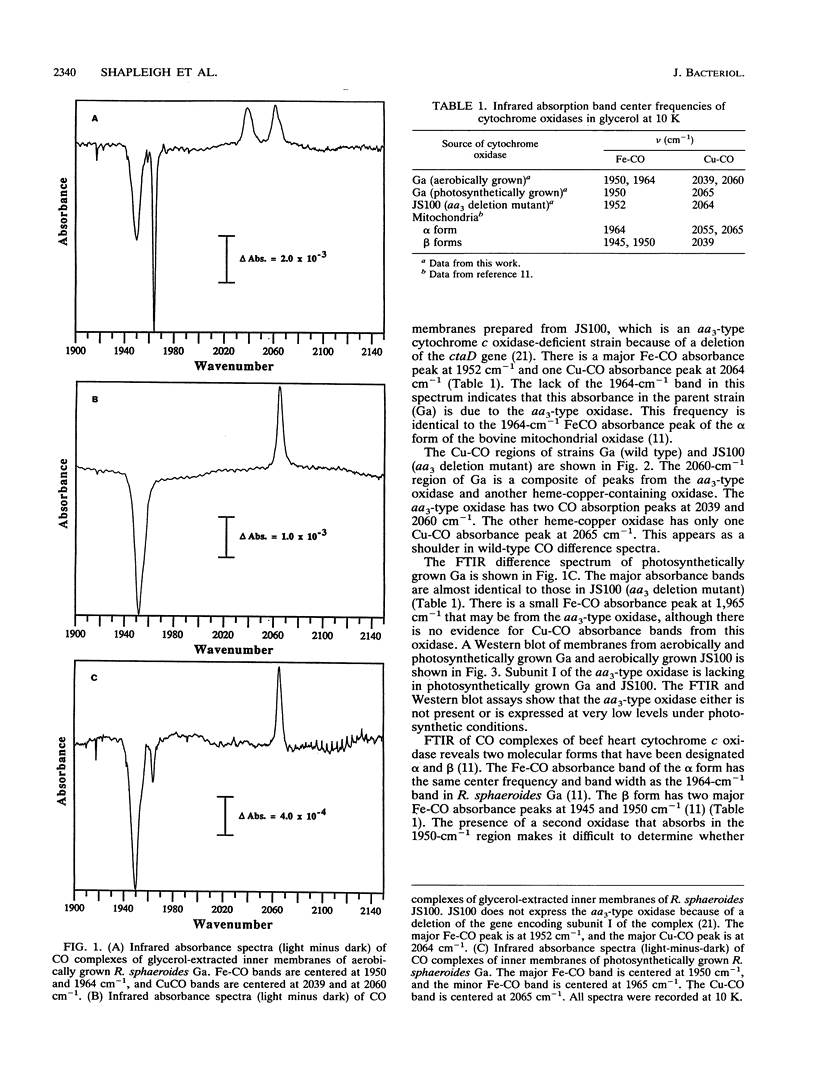
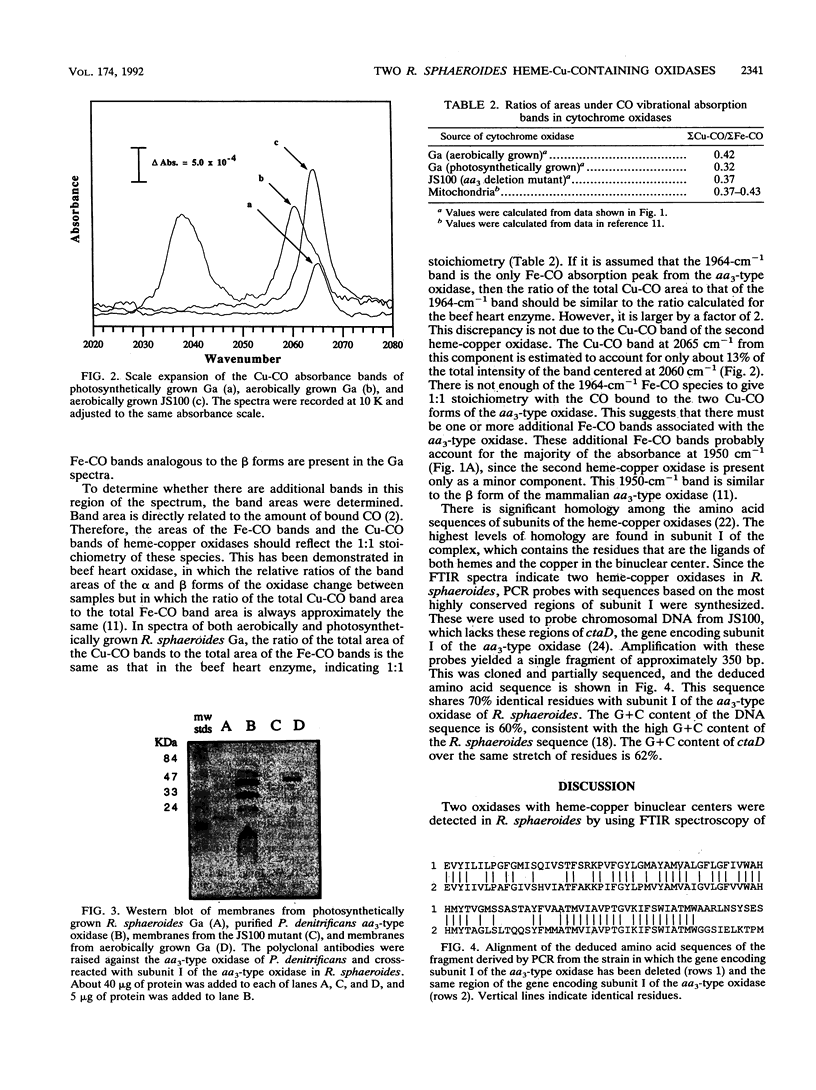
It has recently become evident that many bacterial respiratory oxidases are members of a superfamily that is related to the eukaryotic cytochrome c oxidase. These oxidases catalyze the reduction of oxygen to water at a heme-copper binuclear center. Fourier transform infrared (FTIR) spectroscopy has been used to examine the heme-copper-containing respiratory oxidases of Rhodobacter sphaeroides Ga. This technique monitors the stretching frequency of CO bound at the oxygen binding site and can be used to characterize the oxidases in situ with membrane preparations. Oxidases that have a heme-copper binuclear center are recognizable by FTIR spectroscopy because the bound CO moves from the heme iron to the nearby copper upon photolysis at low temperature, where it exhibits a diagnostic spectrum. The FTIR spectra indicate that the binuclear center of the R. sphaeroides aa3-type cytochrome c oxidase is remarkably similar to that of the bovine mitochondrial oxidase. Upon deletion of the ctaD gene, encoding subunit I of the aa3-type oxidase, substantial cytochrome c oxidase remains in the membranes of aerobically grown R. sphaeroides. This correlates with a second wild-type R. sphaeroides is grown photosynthetically, the chromatophore membranes lack the aa3-type oxidase but have this second heme-copper oxidase. Subunit I of the heme-copper oxidase superfamily contains the binuclear center. Amino acid sequence alignments show that this subunit is structurally very highly conserved among both eukaryotic and prokaryotic species. The polymerase chain reaction was used to show that the chromosome of R. sphaeroides contains at least one other gene that is a homolog of ctaD, the gene encoding subunit I of the aa3-type cytochrome c oxidase.(ABSTRACT TRUNCATED AT 250 WORDS)
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- COHEN-BAZIRE G., SISTROM W. R., STANIER R. Y. Kinetic studies of pigment synthesis by non-sulfur purple bacteria. J Cell Physiol. 1957 Feb;49(1):25–68. doi: 10.1002/jcp.1030490104. [DOI] [PubMed] [Google Scholar]
- Cao J., Shapleigh J., Gennis R., Revzin A., Ferguson-Miller S. The gene encoding cytochrome c oxidase subunit II from Rhodobacter sphaeroides; comparison of the deduced amino acid sequence with sequences of corresponding peptides from other species. Gene. 1991 May 15;101(1):133–137. doi: 10.1016/0378-1119(91)90235-4. [DOI] [PubMed] [Google Scholar]
- Capaldi R. A. Structure and assembly of cytochrome c oxidase. Arch Biochem Biophys. 1990 Aug 1;280(2):252–262. doi: 10.1016/0003-9861(90)90327-u. [DOI] [PubMed] [Google Scholar]
- Capaldi R. A. Structure and function of cytochrome c oxidase. Annu Rev Biochem. 1990;59:569–596. doi: 10.1146/annurev.bi.59.070190.003033. [DOI] [PubMed] [Google Scholar]
- Chan S. I., Li P. M. Cytochrome c oxidase: understanding nature's design of a proton pump. Biochemistry. 1990 Jan 9;29(1):1–12. doi: 10.1021/bi00453a001. [DOI] [PubMed] [Google Scholar]
- Chepuri V., Lemieux L., Au D. C., Gennis R. B. The sequence of the cyo operon indicates substantial structural similarities between the cytochrome o ubiquinol oxidase of Escherichia coli and the aa3-type family of cytochrome c oxidases. J Biol Chem. 1990 Jul 5;265(19):11185–11192. [PubMed] [Google Scholar]
- Chepuri V., Lemieux L., Hill J., Alben J. O., Gennis R. B. Recent studies of the cytochrome o terminal oxidase complex of Escherichia coli. Biochim Biophys Acta. 1990 Jul 25;1018(2-3):124–127. doi: 10.1016/0005-2728(90)90231-r. [DOI] [PubMed] [Google Scholar]
- Einarsdóttir O., Killough P. M., Fee J. A., Woodruff W. H. An infrared study of the binding and photodissociation of carbon monoxide in cytochrome ba3 from Thermus thermophilus. J Biol Chem. 1989 Feb 15;264(5):2405–2408. [PubMed] [Google Scholar]
- Fiamingo F. G., Alben J. O. Structures of photolyzed carboxymyoglobin. Biochemistry. 1985 Dec 31;24(27):7964–7970. doi: 10.1021/bi00348a019. [DOI] [PubMed] [Google Scholar]
- Fiamingo F. G., Altschuld R. A., Moh P. P., Alben J. O. Dynamic interactions of CO with a3Fe and CuB in cytochrome c oxidase in beef heart mitochondria studied by Fourier transform infrared spectroscopy at low temperatures. J Biol Chem. 1982 Feb 25;257(4):1639–1650. [PubMed] [Google Scholar]
- Fiamingo F. G., Jung D. W., Alben J. O. Structural perturbation of the a3-CuB site in mitochondrial cytochrome c oxidase by alcohol solvents. Biochemistry. 1990 May 15;29(19):4627–4633. doi: 10.1021/bi00471a018. [DOI] [PubMed] [Google Scholar]
- Garcia-Horsman J. A., Barquera B., Escamilla J. E. Two different aa3-type cytochromes can be purified from the bacterium Bacillus cereus. Eur J Biochem. 1991 Aug 1;199(3):761–768. doi: 10.1111/j.1432-1033.1991.tb16181.x. [DOI] [PubMed] [Google Scholar]
- Gennis R. B. Some recent advances relating to prokaryotic cytochrome c reductases and cytochrome c oxidases. Biochim Biophys Acta. 1991 May 23;1058(1):21–24. doi: 10.1016/s0005-2728(05)80260-9. [DOI] [PubMed] [Google Scholar]
- Puustinen A., Wikström M. The heme groups of cytochrome o from Escherichia coli. Proc Natl Acad Sci U S A. 1991 Jul 15;88(14):6122–6126. doi: 10.1073/pnas.88.14.6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raitio M., Pispa J. M., Metso T., Saraste M. Are there isoenzymes of cytochrome c oxidase in Paracoccus denitrificans? FEBS Lett. 1990 Feb 26;261(2):431–435. doi: 10.1016/0014-5793(90)80609-m. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste M. Structural features of cytochrome oxidase. Q Rev Biophys. 1990 Nov;23(4):331–366. doi: 10.1017/s0033583500005588. [DOI] [PubMed] [Google Scholar]
- Sasaki T., Motokawa Y., Kikuchi G. Occurrence of both a-type and o-type cytochromes as the functional terminal oxidases in Rhodopseudomonas spheroides. Biochim Biophys Acta. 1970 Mar 3;197(2):284–291. doi: 10.1016/0005-2728(70)90039-3. [DOI] [PubMed] [Google Scholar]
- Sone N., Kutoh E., Sato K. A cytochrome o-type oxidase of the thermophilic bacterium PS3 grown under air-limited conditions. J Biochem. 1990 Apr;107(4):597–602. doi: 10.1093/oxfordjournals.jbchem.a123092. [DOI] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Oyaizu Y., Oyaizu H., Olsen G. J., Woese C. R. Mitochondrial origins. Proc Natl Acad Sci U S A. 1985 Jul;82(13):4443–4447. doi: 10.1073/pnas.82.13.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa S., Caughey W. S. Infrared evidence of cyanide binding to iron and copper sites in bovine heart cytochrome c oxidase. Implications regarding oxygen reduction. J Biol Chem. 1990 May 15;265(14):7945–7958. [PubMed] [Google Scholar]
- Yun C. H., Beci R., Crofts A. R., Kaplan S., Gennis R. B. Cloning and DNA sequencing of the fbc operon encoding the cytochrome bc1 complex from Rhodobacter sphaeroides. Characterization of fbc deletion mutants and complementation by a site-specific mutational variant. Eur J Biochem. 1990 Dec 12;194(2):399–411. doi: 10.1111/j.1432-1033.1990.tb15633.x. [DOI] [PubMed] [Google Scholar]
- van der Oost J., von Wachenfeld C., Hederstedt L., Saraste M. Bacillus subtilis cytochrome oxidase mutants: biochemical analysis and genetic evidence for two aa3-type oxidases. Mol Microbiol. 1991 Aug;5(8):2063–2072. doi: 10.1111/j.1365-2958.1991.tb00829.x. [DOI] [PubMed] [Google Scholar]