Abstract
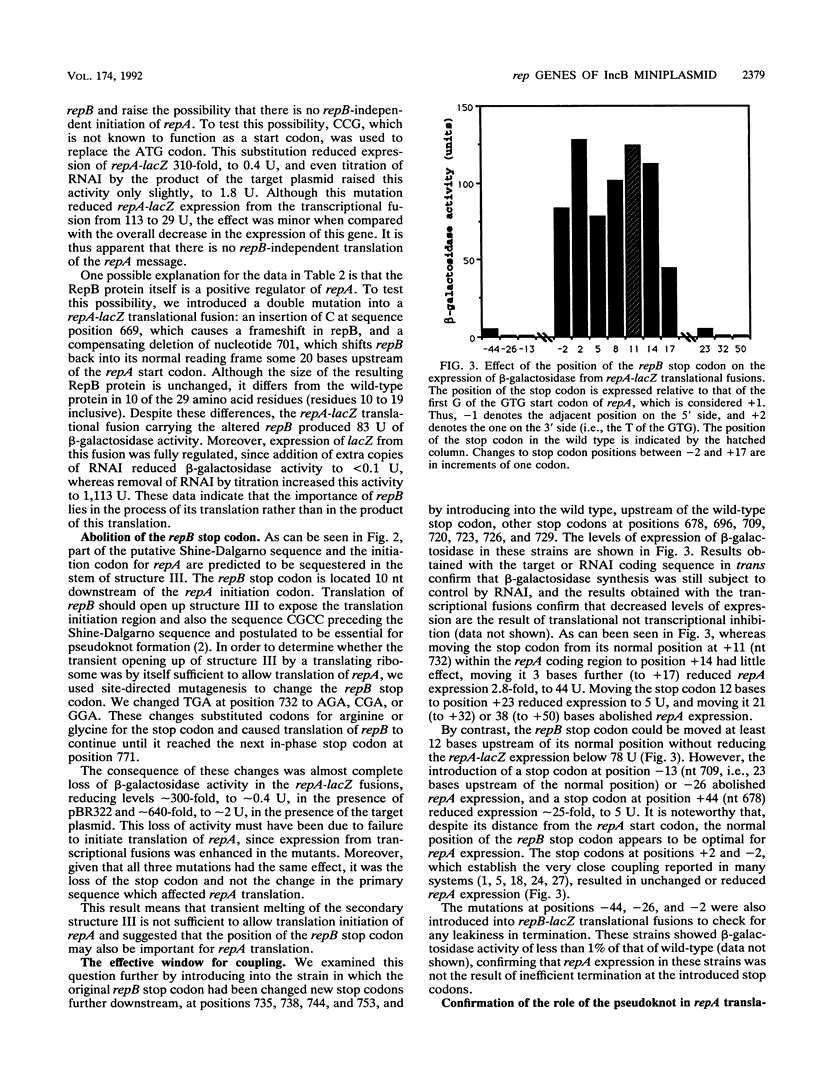
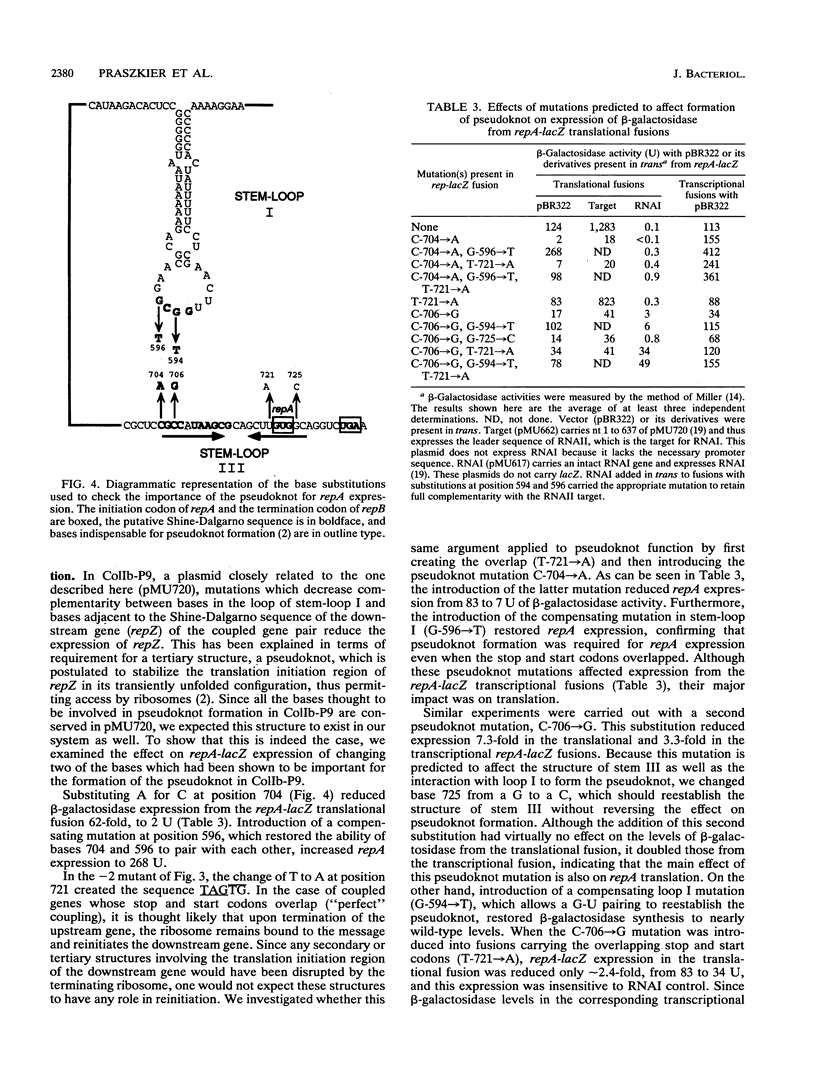
The nature of translational coupling between repB and repA, the overlapping rep genes of the IncB plasmid pMU720, was examined. Mutations in the start codon of the promoter proximal gene, repB, reduced the efficiency of translation of both rep genes. Moreover, there was no independent initiation of repA translation in the absence of repB translation. The position of the repB stop codon was crucial for the efficient expression of repA, with the wild-type positioning being optimal. Translational coupling was found to be totally dependent on the formation of a pseudoknot structure. A model which invokes formation of a pseudoknot to facilitate initiation of repA is proposed.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aksoy S., Squires C. L., Squires C. Translational coupling of the trpB and trpA genes in the Escherichia coli tryptophan operon. J Bacteriol. 1984 Feb;157(2):363–367. doi: 10.1128/jb.157.2.363-367.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K., Kato A., Moriwaki H., Hama C., Shiba K., Mizobuchi K. Positive and negative regulations of plasmid CoLIb-P9 repZ gene expression at the translational level. J Biol Chem. 1991 Feb 25;266(6):3774–3781. [PubMed] [Google Scholar]
- Bird P. I., Pittard J. Demonstration of a third incompatibility function on plasmids already incompatible with group P and group I plasmids. Plasmid. 1983 Mar;9(2):191–200. doi: 10.1016/0147-619x(83)90020-3. [DOI] [PubMed] [Google Scholar]
- Blomberg P., Wagner E. G., Nordström K. Control of replication of plasmid R1: the duplex between the antisense RNA, CopA, and its target, CopT, is processed specifically in vivo and in vitro by RNase III. EMBO J. 1990 Jul;9(7):2331–2340. doi: 10.1002/j.1460-2075.1990.tb07405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A., Yanofsky C. A ribosome binding site sequence is necessary for efficient expression of the distal gene of a translationally-coupled gene pair. Nucleic Acids Res. 1984 Jun 11;12(11):4757–4768. doi: 10.1093/nar/12.11.4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hama C., Takizawa T., Moriwaki H., Mizobuchi K. Role of leader peptide synthesis in repZ gene expression of the ColIb-P9 plasmid. J Biol Chem. 1990 Jun 25;265(18):10666–10673. [PubMed] [Google Scholar]
- Jaeger J. A., Turner D. H., Zuker M. Improved predictions of secondary structures for RNA. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger J. A., Turner D. H., Zuker M. Predicting optimal and suboptimal secondary structure for RNA. Methods Enzymol. 1990;183:281–306. doi: 10.1016/0076-6879(90)83019-6. [DOI] [PubMed] [Google Scholar]
- Kieny M. P., Lathe R., Lecocq J. P. New versatile cloning and sequencing vectors based on bacteriophage M13. Gene. 1983 Dec;26(1):91–99. doi: 10.1016/0378-1119(83)90039-2. [DOI] [PubMed] [Google Scholar]
- Kumar C. C., Novick R. P. Plasmid pT181 replication is regulated by two countertranscripts. Proc Natl Acad Sci U S A. 1985 Feb;82(3):638–642. doi: 10.1073/pnas.82.3.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light J., Molin S. Post-transcriptional control of expression of the repA gene of plasmid R1 mediated by a small RNA molecule. EMBO J. 1983;2(1):93–98. doi: 10.1002/j.1460-2075.1983.tb01387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MONOD J., COHEN-BAZIRE G., COHN M. Sur la biosynthèse de la beta-galactosidase (lactase) chez Escherichia coli; la spécificité de l'induction. Biochim Biophys Acta. 1951 Nov;7(4):585–599. doi: 10.1016/0006-3002(51)90072-8. [DOI] [PubMed] [Google Scholar]
- Masukata H., Tomizawa J. Control of primer formation for ColE1 plasmid replication: conformational change of the primer transcript. Cell. 1986 Jan 17;44(1):125–136. doi: 10.1016/0092-8674(86)90491-5. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Nikoletti S., Bird P., Praszkier J., Pittard J. Analysis of the incompatibility determinants of I-complex plasmids. J Bacteriol. 1988 Mar;170(3):1311–1318. doi: 10.1128/jb.170.3.1311-1318.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P., Iordanescu S., Projan S. J., Kornblum J., Edelman I. pT181 plasmid replication is regulated by a countertranscript-driven transcriptional attenuator. Cell. 1989 Oct 20;59(2):395–404. doi: 10.1016/0092-8674(89)90300-0. [DOI] [PubMed] [Google Scholar]
- Oppenheim D. S., Yanofsky C. Translational coupling during expression of the tryptophan operon of Escherichia coli. Genetics. 1980 Aug;95(4):785–795. doi: 10.1093/genetics/95.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praszkier J., Bird P., Nikoletti S., Pittard J. Role of countertranscript RNA in the copy number control system of an IncB miniplasmid. J Bacteriol. 1989 Sep;171(9):5056–5064. doi: 10.1128/jb.171.9.5056-5064.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praszkier J., Wei T., Siemering K., Pittard J. Comparative analysis of the replication regions of IncB, IncK, and IncZ plasmids. J Bacteriol. 1991 Apr;173(7):2393–2397. doi: 10.1128/jb.173.7.2393-2397.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa M. D. Structure analysis of three T7 late mRNA ribosome binding sites. J Mol Biol. 1981 Mar 25;147(1):55–71. doi: 10.1016/0022-2836(81)90079-6. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schümperli D., McKenney K., Sobieski D. A., Rosenberg M. Translational coupling at an intercistronic boundary of the Escherichia coli galactose operon. Cell. 1982 Oct;30(3):865–871. doi: 10.1016/0092-8674(82)90291-4. [DOI] [PubMed] [Google Scholar]
- Steitz J. A. Polypeptide chain initiation: nucleotide sequences of the three ribosomal binding sites in bacteriophage R17 RNA. Nature. 1969 Dec 6;224(5223):957–964. doi: 10.1038/224957a0. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Theisen M., Neuhard J. Translational coupling in the pyrF operon of Salmonella typhimurium. Mol Gen Genet. 1990 Jul;222(2-3):345–352. doi: 10.1007/BF00633839. [DOI] [PubMed] [Google Scholar]
- Tomizawa J. Control of ColE1 plasmid replication: binding of RNA I to RNA II and inhibition of primer formation. Cell. 1986 Oct 10;47(1):89–97. doi: 10.1016/0092-8674(86)90369-7. [DOI] [PubMed] [Google Scholar]
- Tomizawa J. Control of ColE1 plasmid replication: the process of binding of RNA I to the primer transcript. Cell. 1984 Oct;38(3):861–870. doi: 10.1016/0092-8674(84)90281-2. [DOI] [PubMed] [Google Scholar]
- Tomizawa J., Itoh T., Selzer G., Som T. Inhibition of ColE1 RNA primer formation by a plasmid-specified small RNA. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1421–1425. doi: 10.1073/pnas.78.3.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandeyar M. A., Weiner M. P., Hutton C. J., Batt C. A. A simple and rapid method for the selection of oligodeoxynucleotide-directed mutants. Gene. 1988 May 15;65(1):129–133. doi: 10.1016/0378-1119(88)90425-8. [DOI] [PubMed] [Google Scholar]
- Yang J., Pittard J. Molecular analysis of the regulatory region of the Escherichia coli K-12 tyrB gene. J Bacteriol. 1987 Oct;169(10):4710–4715. doi: 10.1128/jb.169.10.4710-4715.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989 Apr 7;244(4900):48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]


