Abstract
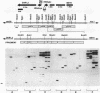
Expression of allantoin pathway genes is subject to induction and nitrogen catabolite repression. Two classes of cis-dominant mutations (DUR80 and DUR1,2-Oh) result in overproduction of DUR1,2 mRNA. In DUR80 mutants, DUR1,2 expression remained inducible, nitrogen catabolite repression sensitive, and unresponsive to cell ploidy, i.e., overproduction was superimposed on normal gene regulation. DUR1,2-Oh mutations, in contrast, generated a pattern of DUR1,2 expression similar to that often reported when a Ty element inserts upstream of a gene, the ROAM phenotype. We analyzed four independent DUR80 and DUR1,2-Oh alleles. The DUR1,2-Oh mutation was, as expected, a Ty insertion at -445 3' of the native DUR1,2 upstream activation sequences (UASs). All three DUR80 alleles were also Ty insertions between -644 and -653 immediately 5' of the native DUR1,2 USASs. We suggest that the difference in DUR1,2-Oh and DUR80 phenotypes depends on whether the native cis-acting elements and transcription factors associated with them can operate. If they can, enhancement of normally regulated DUR1,2 expression is observed. This is a novel phenotype for Ty insertions. If the native DUR1,2 cis-acting elements are not present, the case when Ty insertion occurs 3' of them, a ROAM phenotype is generated. Nitrogen-regulated upstream activation sequence (UASNTR)-homologous sequences present in the Ty delta elements rather than cis-acting elements required for Ty transcription are the most likely candidates to serve as the cis-acting elements mediating the DUR80 phenotype.
Full text
PDF

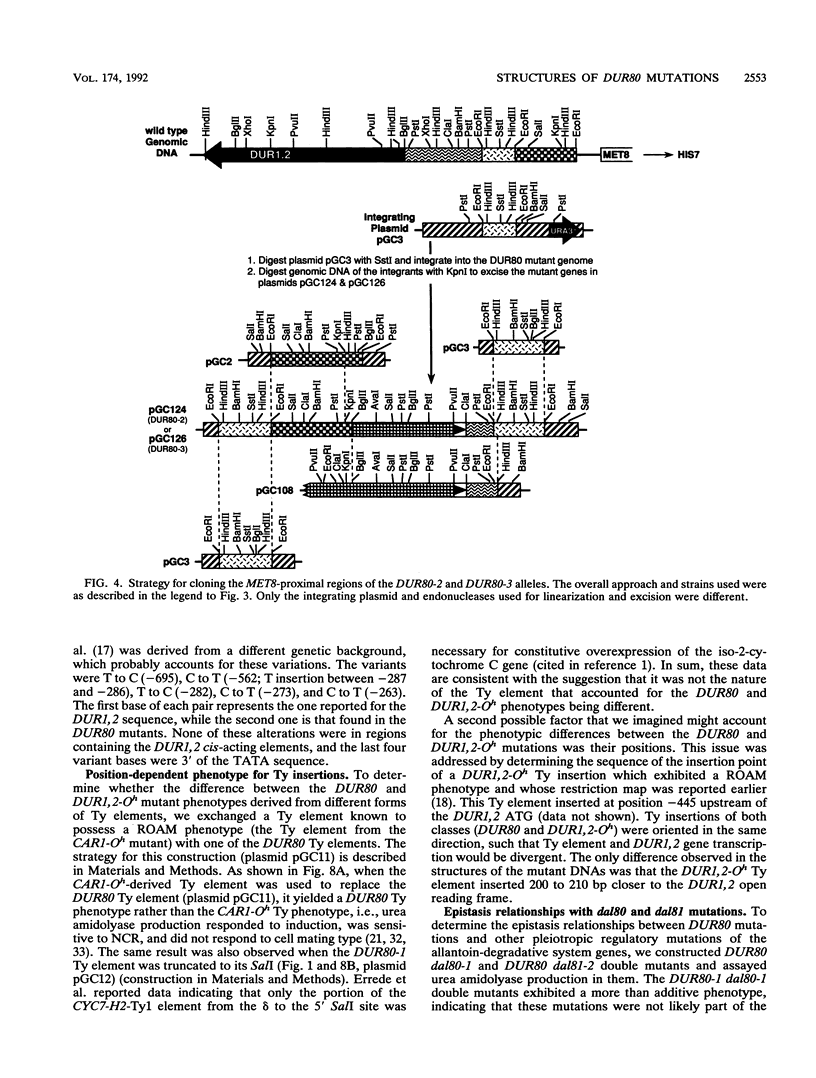
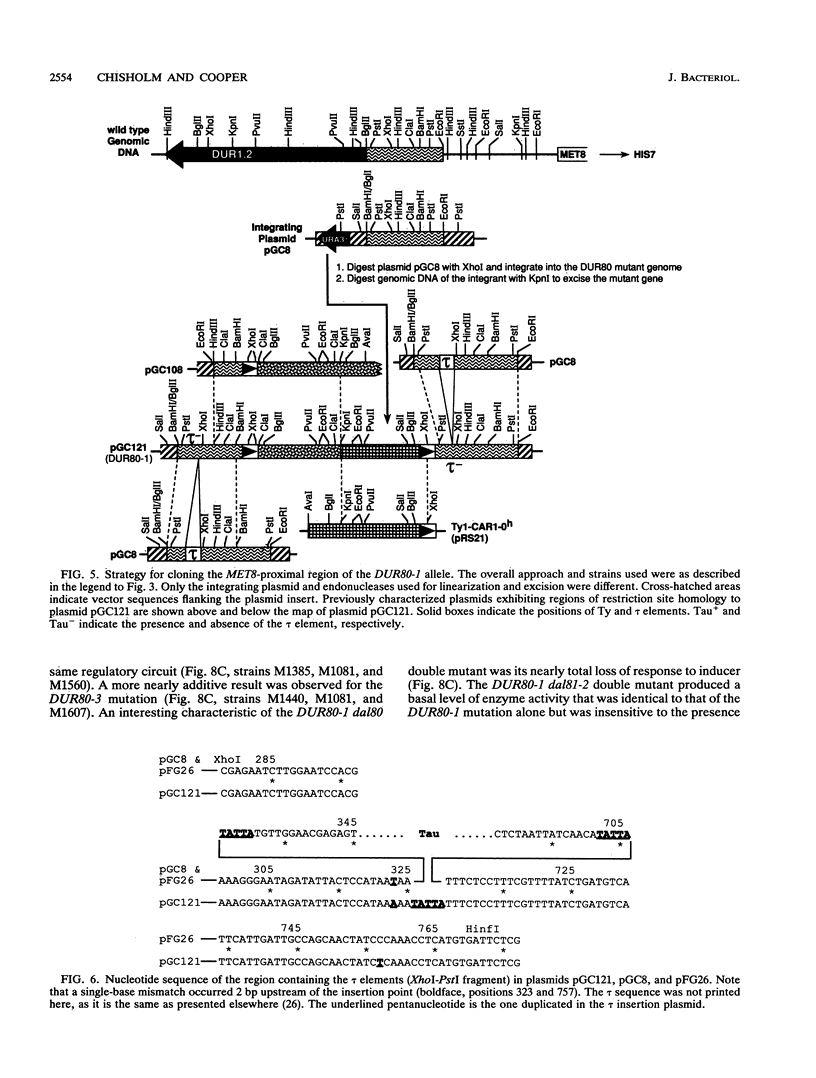
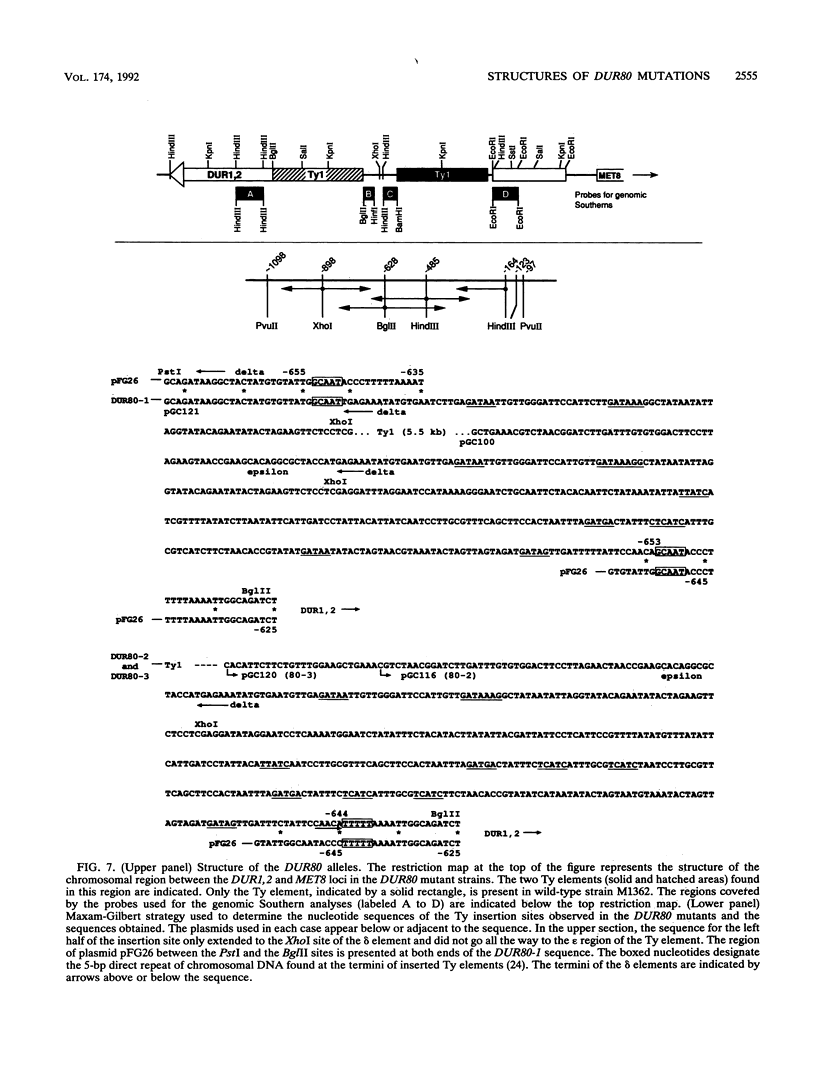
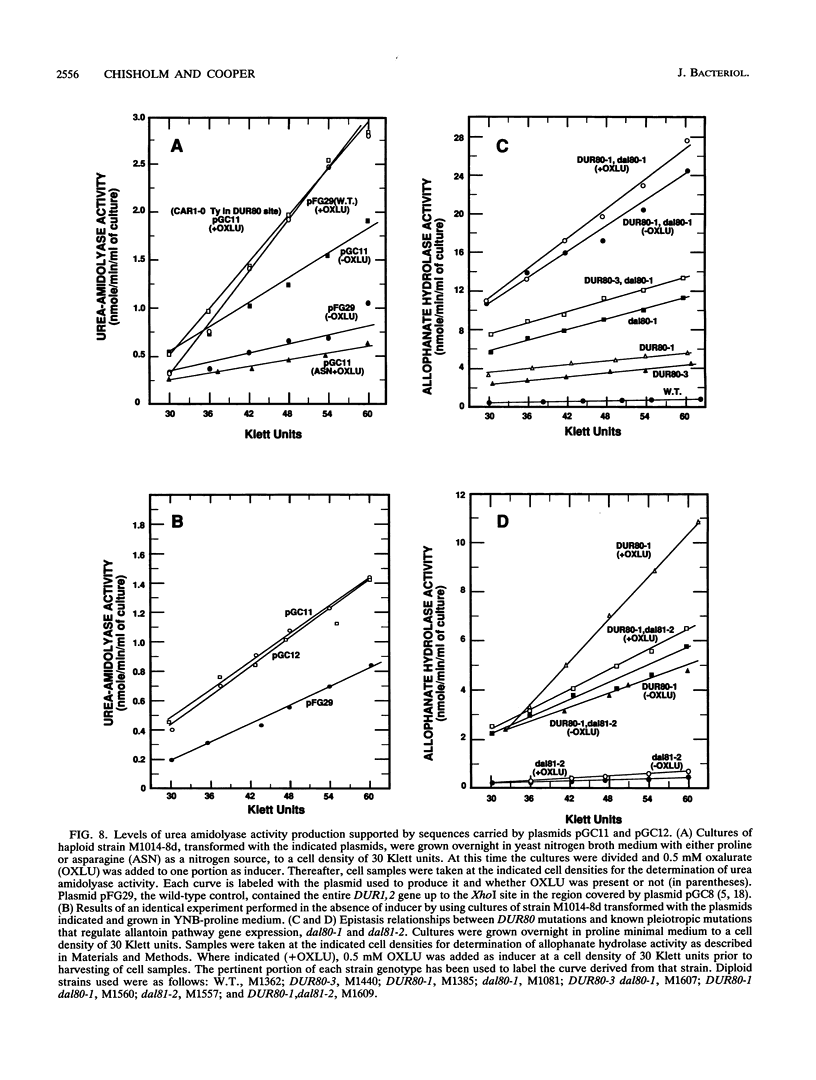


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bricmont P. A., Cooper T. G. A gene product needed for induction of allantoin system genes in Saccharomyces cerevisiae but not for their transcriptional activation. Mol Cell Biol. 1989 Sep;9(9):3869–3877. doi: 10.1128/mcb.9.9.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricmont P. A., Daugherty J. R., Cooper T. G. The DAL81 gene product is required for induced expression of two differently regulated nitrogen catabolic genes in Saccharomyces cerevisiae. Mol Cell Biol. 1991 Feb;11(2):1161–1166. doi: 10.1128/mcb.11.2.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bysani N., Daugherty J. R., Cooper T. G. Saturation mutagenesis of the UASNTR (GATAA) responsible for nitrogen catabolite repression-sensitive transcriptional activation of the allantoin pathway genes in Saccharomyces cerevisiae. J Bacteriol. 1991 Aug;173(16):4977–4982. doi: 10.1128/jb.173.16.4977-4982.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm G. E., Genbauffe F. S., Cooper T. G. tau, a repeated DNA sequence in yeast. Proc Natl Acad Sci U S A. 1984 May;81(10):2965–2969. doi: 10.1073/pnas.81.10.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm G., Cooper T. G. Isolation and characterization of mutants that produce the allantoin-degrading enzymes constitutively in Saccharomyces cerevisiae. Mol Cell Biol. 1982 Sep;2(9):1088–1095. doi: 10.1128/mcb.2.9.1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm G., Cooper T. cis-Dominant mutations which dramatically enhance DUR1,2 gene expression without affecting its normal regulation. Mol Cell Biol. 1984 May;4(5):947–955. doi: 10.1128/mcb.4.5.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm V. T., Lea H. Z., Rai R., Cooper T. G. Regulation of allantoate transport in wild-type and mutant strains of Saccharomyces cerevisiae. J Bacteriol. 1987 Apr;169(4):1684–1690. doi: 10.1128/jb.169.4.1684-1690.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T. G. Allantoin degradation by Saccharomyces cerevisiae--a model system for gene regulation and metabolic integration. Adv Enzymol Relat Areas Mol Biol. 1984;56:91–139. doi: 10.1002/9780470123027.ch2. [DOI] [PubMed] [Google Scholar]
- Cooper T. G., Chisholm V. T., Cho H. J., Yoo H. S. Allantoin transport in Saccharomyces cerevisiae is regulated by two induction systems. J Bacteriol. 1987 Oct;169(10):4660–4667. doi: 10.1128/jb.169.10.4660-4667.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T. G., Ferguson D., Rai R., Bysani N. The GLN3 gene product is required for transcriptional activation of allantoin system gene expression in Saccharomyces cerevisiae. J Bacteriol. 1990 Feb;172(2):1014–1018. doi: 10.1128/jb.172.2.1014-1018.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T. G., Lam C., Turoscy V. Structural analysis of the dur loci in S. cerevisiae: two domains of a single multifunctional gene. Genetics. 1980 Mar;94(3):555–580. doi: 10.1093/genetics/94.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T. G. Mutants of Saccharomyces cerevisiae possessing fully induced levels of urea amido-lyase in the absence of added inducer. Biochem Biophys Res Commun. 1978 Jun 29;82(4):1258–1263. doi: 10.1016/0006-291x(78)90323-6. [DOI] [PubMed] [Google Scholar]
- Cooper T. G., Rai R., Yoo H. S. Requirement of upstream activation sequences for nitrogen catabolite repression of the allantoin system genes in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Dec;9(12):5440–5444. doi: 10.1128/mcb.9.12.5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T. G., Sumrada R. A. What is the function of nitrogen catabolite repression in Saccharomyces cerevisiae? J Bacteriol. 1983 Aug;155(2):623–627. doi: 10.1128/jb.155.2.623-627.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham T. S., Cooper T. G. Expression of the DAL80 gene, whose product is homologous to the GATA factors and is a negative regulator of multiple nitrogen catabolic genes in Saccharomyces cerevisiae, is sensitive to nitrogen catabolite repression. Mol Cell Biol. 1991 Dec;11(12):6205–6215. doi: 10.1128/mcb.11.12.6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois E., Jacobs E., Jauniaux J. C. Expression of the ROAM mutations in Saccharomyces cerevisiae: involvement of trans-acting regulatory elements and relation with the Ty1 transcription. EMBO J. 1982;1(9):1133–1139. doi: 10.1002/j.1460-2075.1982.tb01308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genbauffe F. S., Chisholm G. E., Cooper T. G. Tau, sigma, and delta. A family of repeated elements in yeast. J Biol Chem. 1984 Aug 25;259(16):10518–10525. [PubMed] [Google Scholar]
- Genbauffe F. S., Cooper T. G. Induction and repression of the urea amidolyase gene in Saccharomyces cerevisiae. Mol Cell Biol. 1986 Nov;6(11):3954–3964. doi: 10.1128/mcb.6.11.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genbauffe F. S., Cooper T. G. The urea amidolyase (DUR1,2) gene of Saccharomyces cerevisiae. DNA Seq. 1991;2(1):19–32. doi: 10.3109/10425179109008435. [DOI] [PubMed] [Google Scholar]
- Herbomel P. Synergistic activation of eukaryotic transcription: the multiacceptor target hypothesis. New Biol. 1990 Dec;2(12):1063–1070. [PubMed] [Google Scholar]
- Jauniaux J. C., Dubois E., Vissers S., Crabeel M., Wiame J. M. Molecular cloning, DNA structure, and RNA analysis of the arginase gene in Saccharomyces cerevisiae. A study of cis-dominant regulatory mutations. EMBO J. 1982;1(9):1125–1131. doi: 10.1002/j.1460-2075.1982.tb01307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovari L., Sumrada R., Kovari I., Cooper T. G. Multiple positive and negative cis-acting elements mediate induced arginase (CAR1) gene expression in Saccharomyces cerevisiae. Mol Cell Biol. 1990 Oct;10(10):5087–5097. doi: 10.1128/mcb.10.10.5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawther R. P., Cooper T. G. Kinetics of induced and repressed enzyme synthesis in Saccharomyces cerevisiae. J Bacteriol. 1975 Mar;121(3):1064–1073. doi: 10.1128/jb.121.3.1064-1073.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoine Y., Dubois E., Wiame J. M. The regulation of urea amidolyase of Saccharomyces cerevisiae: mating type influence on a constitutivity mutation acting in cis. Mol Gen Genet. 1978 Nov 9;166(3):251–258. [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Olive M. G., Daugherty J. R., Cooper T. G. DAL82, a second gene required for induction of allantoin system gene transcription in Saccharomyces cerevisiae. J Bacteriol. 1991 Jan;173(1):255–261. doi: 10.1128/jb.173.1.255-261.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R., Genbauffe F. S., Cooper T. G. Structure and transcription of the allantoate permease gene (DAL5) from Saccharomyces cerevisiae. J Bacteriol. 1988 Jan;170(1):266–271. doi: 10.1128/jb.170.1.266-271.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R., Genbauffe F., Lea H. Z., Cooper T. G. Transcriptional regulation of the DAL5 gene in Saccharomyces cerevisiae. J Bacteriol. 1987 Aug;169(8):3521–3524. doi: 10.1128/jb.169.8.3521-3524.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumrada R. A., Cooper T. G. Isolation of the CAR1 gene from Saccharomyces cerevisiae and analysis of its expression. Mol Cell Biol. 1982 Dec;2(12):1514–1523. doi: 10.1128/mcb.2.12.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumrada R. A., Cooper T. G. Point mutation generates constitutive expression of an inducible eukaryotic gene. Proc Natl Acad Sci U S A. 1985 Feb;82(3):643–647. doi: 10.1073/pnas.82.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumrada R., Cooper T. G. Oxaluric acid: a non-metabolizable inducer of the allantoin degradative enzymes in Saccharomyces cerevisiae. J Bacteriol. 1974 Mar;117(3):1240–1247. doi: 10.1128/jb.117.3.1240-1247.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turoscy V., Cooper T. G. Pleiotropic control of five eucaryotic genes by multiple regulatory elements. J Bacteriol. 1982 Sep;151(3):1237–1246. doi: 10.1128/jb.151.3.1237-1246.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney P. A., Cooper T. G., Magasanik B. The induction of urea carboxylase and allophanate hydrolase in Saccharomyces cerevisiae. J Biol Chem. 1973 Sep 10;248(17):6203–6209. [PubMed] [Google Scholar]
- Wickerham L. J. A Critical Evaluation of the Nitrogen Assimilation Tests Commonly Used in the Classification of Yeasts. J Bacteriol. 1946 Sep;52(3):293–301. [PMC free article] [PubMed] [Google Scholar]
- Yoo H. S., Cooper T. G. The DAL7 promoter consists of multiple elements that cooperatively mediate regulation of the gene's expression. Mol Cell Biol. 1989 Aug;9(8):3231–3243. doi: 10.1128/mcb.9.8.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H. S., Genbauffe F. S., Cooper T. G. Identification of the ureidoglycolate hydrolase gene in the DAL gene cluster of Saccharomyces cerevisiae. Mol Cell Biol. 1985 Sep;5(9):2279–2288. doi: 10.1128/mcb.5.9.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]