Abstract
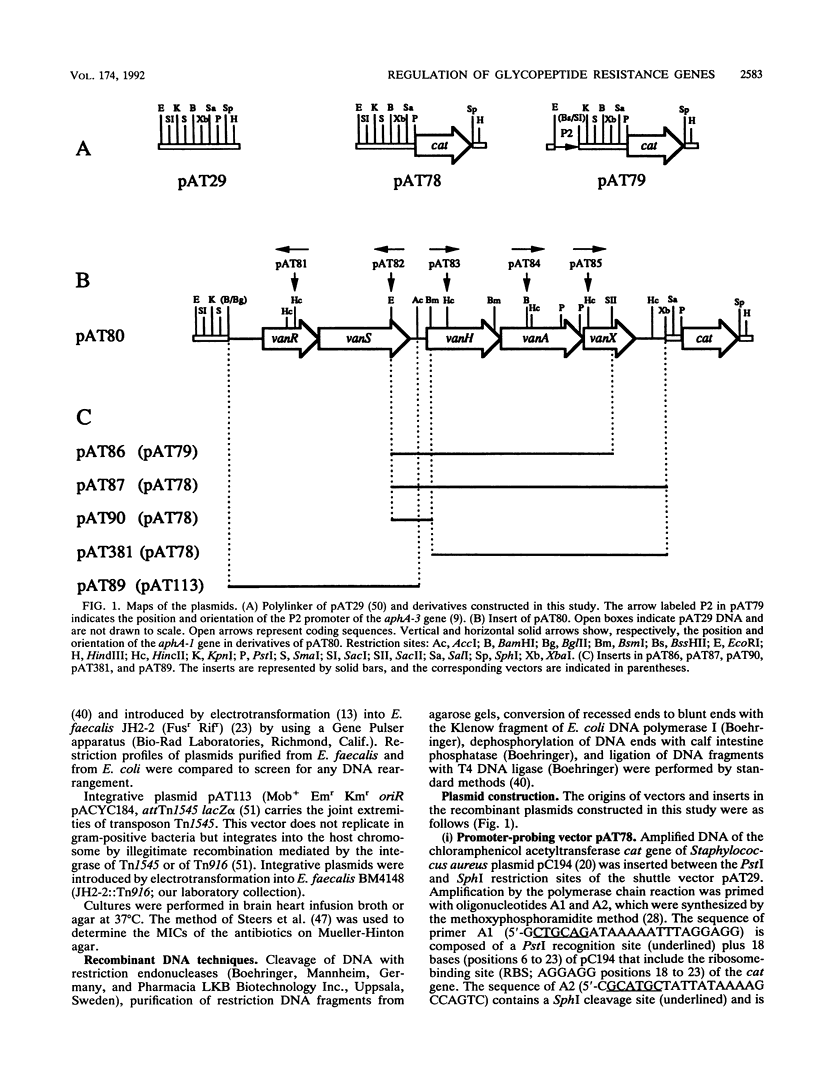
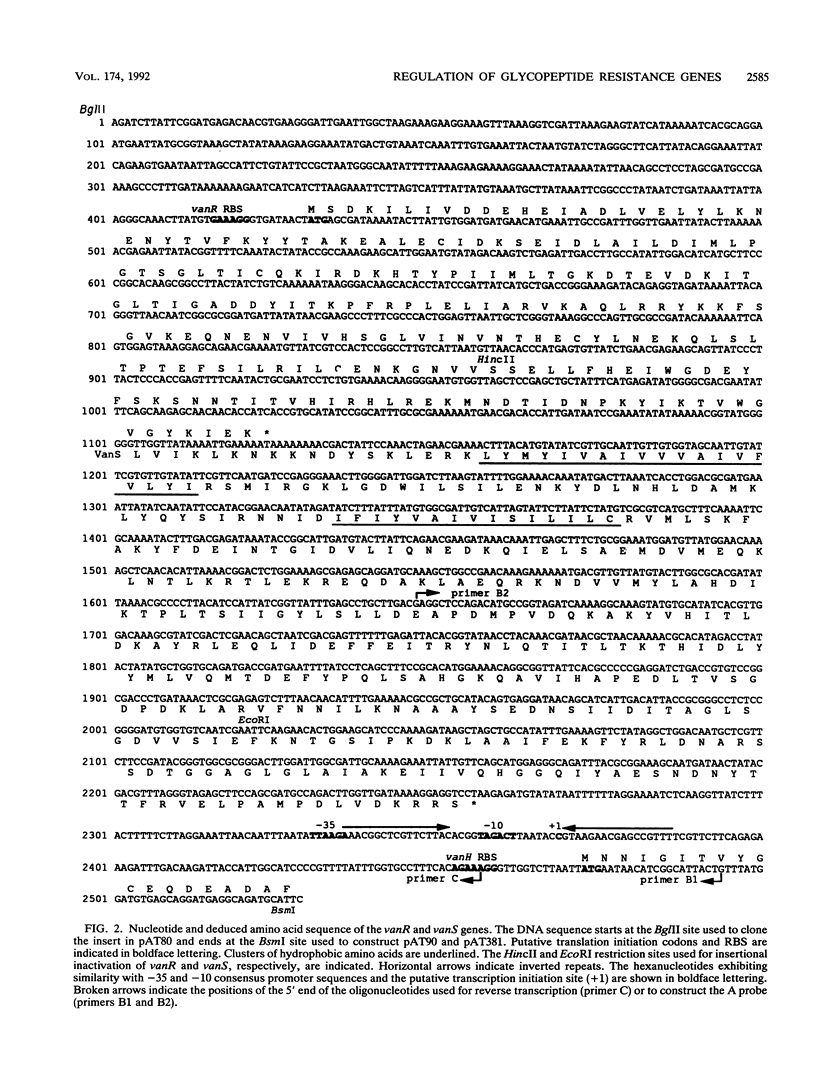
Plasmid pIP816 of Enterococcus faecium BM4147 confers inducible resistance to vancomycin and encodes the VanH dehydrogenase and the VanA ligase for synthesis of depsipeptide-containing peptidoglycan precursors which bind the antibiotic with reduced affinity. We have characterized a cluster of five genes of pIP816 sufficient for peptidoglycan synthesis in the presence of vancomycin. The distal part of the van cluster encodes VanH, VanA, and a third enzyme, VanX, all of which are necessary for resistance. Synthesis of these enzymes was regulated at the transcriptional level by the VanS-VanR two-component regulatory system encoded by the proximal part of the cluster. VanR was a transcriptional activator related to response regulators of the OmpR subclass. VanS stimulated VanR-dependent transcription and was related to membrane-associated histidine protein kinases which control the level of phosphorylation of response regulators. Analysis of transcriptional fusions with a reporter gene and RNA mapping indicated that the VanR-VanS two-component regulatory system activates a promoter used for cotranscription of the vanH, vanA, and vanX resistance genes.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambulos N. P., Jr, Chow J. H., Mongkolsuk S., Preis L. H., Vollmar W. R., 2nd, Lovett P. S. Constitutive variants of the pC194 cat gene exhibit DNA alterations in the vicinity of the ribosome binding site sequence. Gene. 1984 May;28(2):171–176. doi: 10.1016/0378-1119(84)90254-3. [DOI] [PubMed] [Google Scholar]
- Arthur M., Molinas C., Dutka-Malen S., Courvalin P. Structural relationship between the vancomycin resistance protein VanH and 2-hydroxycarboxylic acid dehydrogenases. Gene. 1991 Jul 15;103(1):133–134. doi: 10.1016/0378-1119(91)90405-z. [DOI] [PubMed] [Google Scholar]
- Brisson-Noël A., Dutka-Malen S., Molinas C., Leclercq R., Courvalin P. Cloning and heterospecific expression of the resistance determinant vanA encoding high-level resistance to glycopeptides in Enterococcus faecium BM4147. Antimicrob Agents Chemother. 1990 May;34(5):924–927. doi: 10.1128/aac.34.5.924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugg T. D., Dutka-Malen S., Arthur M., Courvalin P., Walsh C. T. Identification of vancomycin resistance protein VanA as a D-alanine:D-alanine ligase of altered substrate specificity. Biochemistry. 1991 Feb 26;30(8):2017–2021. doi: 10.1021/bi00222a002. [DOI] [PubMed] [Google Scholar]
- Bugg T. D., Wright G. D., Dutka-Malen S., Arthur M., Courvalin P., Walsh C. T. Molecular basis for vancomycin resistance in Enterococcus faecium BM4147: biosynthesis of a depsipeptide peptidoglycan precursor by vancomycin resistance proteins VanH and VanA. Biochemistry. 1991 Oct 29;30(43):10408–10415. doi: 10.1021/bi00107a007. [DOI] [PubMed] [Google Scholar]
- Caillaud F., Trieu-Cuot P., Carlier C., Courvalin P. Nucleotide sequence of the kanamycin resistance determinant of the pneumococcal transposon Tn1545: evolutionary relationships and transcriptional analysis of aphA-3 genes. Mol Gen Genet. 1987 May;207(2-3):509–513. doi: 10.1007/BF00331623. [DOI] [PubMed] [Google Scholar]
- Comeau D. E., Ikenaka K., Tsung K. L., Inouye M. Primary characterization of the protein products of the Escherichia coli ompB locus: structure and regulation of synthesis of the OmpR and EnvZ proteins. J Bacteriol. 1985 Nov;164(2):578–584. doi: 10.1128/jb.164.2.578-584.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courvalin P. M., Shaw W. V., Jacob A. E. Plasmid-mediated mechanisms of resistance to aminoglycoside-aminocyclitol antibiotics and to chloramphenicol in group D streptococci. Antimicrob Agents Chemother. 1978 May;13(5):716–725. doi: 10.1128/aac.13.5.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courvalin P. Resistance of enterococci to glycopeptides. Antimicrob Agents Chemother. 1990 Dec;34(12):2291–2296. doi: 10.1128/aac.34.12.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Rodz A. L., Gilmore M. S. High efficiency introduction of plasmid DNA into glycine treated Enterococcus faecalis by electroporation. Mol Gen Genet. 1990 Oct;224(1):152–154. doi: 10.1007/BF00259462. [DOI] [PubMed] [Google Scholar]
- Dutka-Malen S., Molinas C., Arthur M., Courvalin P. The VANA glycopeptide resistance protein is related to D-alanyl-D-alanine ligase cell wall biosynthesis enzymes. Mol Gen Genet. 1990 Dec;224(3):364–372. doi: 10.1007/BF00262430. [DOI] [PubMed] [Google Scholar]
- Forst S., Comeau D., Norioka S., Inouye M. Localization and membrane topology of EnvZ, a protein involved in osmoregulation of OmpF and OmpC in Escherichia coli. J Biol Chem. 1987 Dec 5;262(34):16433–16438. [PubMed] [Google Scholar]
- Forst S., Delgado J., Ramakrishnan G., Inouye M. Regulation of ompC and ompF expression in Escherichia coli in the absence of envZ. J Bacteriol. 1988 Nov;170(11):5080–5085. doi: 10.1128/jb.170.11.5080-5085.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forst S., Delgado J., Rampersaud A., Inouye M. In vivo phosphorylation of OmpR, the transcription activator of the ompF and ompC genes in Escherichia coli. J Bacteriol. 1990 Jun;172(6):3473–3477. doi: 10.1128/jb.172.6.3473-3477.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatron M. F., Rapoport G. Biosynthesis of the parasporal inclusion of Bacillus thuringiensis: half-life of its corresponding messenger RNA. Biochimie. 1972;54(10):1291–1301. doi: 10.1016/s0300-9084(72)80070-1. [DOI] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J Bacteriol. 1982 May;150(2):815–825. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igo M. M., Ninfa A. J., Silhavy T. J. A bacterial environmental sensor that functions as a protein kinase and stimulates transcriptional activation. Genes Dev. 1989 May;3(5):598–605. doi: 10.1101/gad.3.5.598. [DOI] [PubMed] [Google Scholar]
- Igo M. M., Ninfa A. J., Stock J. B., Silhavy T. J. Phosphorylation and dephosphorylation of a bacterial transcriptional activator by a transmembrane receptor. Genes Dev. 1989 Nov;3(11):1725–1734. doi: 10.1101/gad.3.11.1725. [DOI] [PubMed] [Google Scholar]
- Jacob A. E., Hobbs S. J. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J Bacteriol. 1974 Feb;117(2):360–372. doi: 10.1128/jb.117.2.360-372.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq R., Bingen E., Su Q. H., Lambert-Zechovski N., Courvalin P., Duval J. Effects of combinations of beta-lactams, daptomycin, gentamicin, and glycopeptides against glycopeptide-resistant enterococci. Antimicrob Agents Chemother. 1991 Jan;35(1):92–98. doi: 10.1128/aac.35.1.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq R., Derlot E., Duval J., Courvalin P. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N Engl J Med. 1988 Jul 21;319(3):157–161. doi: 10.1056/NEJM198807213190307. [DOI] [PubMed] [Google Scholar]
- Leonhardt H., Alonso J. C. S1 mapping of the pC194 encoded chloramphenicol acetyltransferase gene in Bacillus subtilis. Nucleic Acids Res. 1988 Feb 25;16(4):1618–1618. doi: 10.1093/nar/16.4.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Mabilat C., Goussard S., Sougakoff W., Spencer R. C., Courvalin P. Direct sequencing of the amplified structural gene and promoter for the extended-broad-spectrum beta-lactamase TEM-9 (RHH-1) of Klebsiella pneumoniae. Plasmid. 1990 Jan;23(1):27–34. doi: 10.1016/0147-619x(90)90041-a. [DOI] [PubMed] [Google Scholar]
- Makino K., Shinagawa H., Amemura M., Nakata A. Nucleotide sequence of the phoB gene, the positive regulatory gene for the phosphate regulon of Escherichia coli K-12. J Mol Biol. 1986 Jul 5;190(1):37–44. doi: 10.1016/0022-2836(86)90073-2. [DOI] [PubMed] [Google Scholar]
- Makino K., Shinagawa H., Amemura M., Nakata A. Nucleotide sequence of the phoR gene, a regulatory gene for the phosphate regulon of Escherichia coli. J Mol Biol. 1986 Dec 5;192(3):549–556. doi: 10.1016/0022-2836(86)90275-5. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Mizuno T., Wurtzel E. T., Inouye M. Osmoregulation of gene expression. II. DNA sequence of the envZ gene of the ompB operon of Escherichia coli and characterization of its gene product. J Biol Chem. 1982 Nov 25;257(22):13692–13698. [PubMed] [Google Scholar]
- Moran C. P., Jr, Lang N., LeGrice S. F., Lee G., Stephens M., Sonenshein A. L., Pero J., Losick R. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol Gen Genet. 1982;186(3):339–346. doi: 10.1007/BF00729452. [DOI] [PubMed] [Google Scholar]
- Nagarajan R. Antibacterial activities and modes of action of vancomycin and related glycopeptides. Antimicrob Agents Chemother. 1991 Apr;35(4):605–609. doi: 10.1128/aac.35.4.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nara F., Matsuyama S., Mizuno T., Mizushima S. Molecular analysis of mutant ompR genes exhibiting different phenotypes as to osmoregulation of the ompF and ompC genes of Escherichia coli. Mol Gen Genet. 1986 Feb;202(2):194–199. doi: 10.1007/BF00331636. [DOI] [PubMed] [Google Scholar]
- Nicas T. I., Wu C. Y., Hobbs J. N., Jr, Preston D. A., Allen N. E. Characterization of vancomycin resistance in Enterococcus faecium and Enterococcus faecalis. Antimicrob Agents Chemother. 1989 Jul;33(7):1121–1124. doi: 10.1128/aac.33.7.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Ronson C. W., Nixon B. T., Ausubel F. M. Conserved domains in bacterial regulatory proteins that respond to environmental stimuli. Cell. 1987 Jun 5;49(5):579–581. doi: 10.1016/0092-8674(87)90530-7. [DOI] [PubMed] [Google Scholar]
- Sanders D. A., Gillece-Castro B. L., Stock A. M., Burlingame A. L., Koshland D. E., Jr Identification of the site of phosphorylation of the chemotaxis response regulator protein, CheY. J Biol Chem. 1989 Dec 25;264(36):21770–21778. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw W. V. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 1975;43:737–755. doi: 10.1016/0076-6879(75)43141-x. [DOI] [PubMed] [Google Scholar]
- Shlaes D. M., Bouvet A., Devine C., Shlaes J. H., al-Obeid S., Williamson R. Inducible, transferable resistance to vancomycin in Enterococcus faecalis A256. Antimicrob Agents Chemother. 1989 Feb;33(2):198–203. doi: 10.1128/aac.33.2.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlaes D. M., Etter L., Gutmann L. Synergistic killing of vancomycin-resistant enterococci of classes A, B, and C by combinations of vancomycin, penicillin, and gentamicin. Antimicrob Agents Chemother. 1991 Apr;35(4):776–779. doi: 10.1128/aac.35.4.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. A new computer method for the storage and manipulation of DNA gel reading data. Nucleic Acids Res. 1980 Aug 25;8(16):3673–3694. doi: 10.1093/nar/8.16.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock A., Koshland D. E., Jr, Stock J. Homologies between the Salmonella typhimurium CheY protein and proteins involved in the regulation of chemotaxis, membrane protein synthesis, and sporulation. Proc Natl Acad Sci U S A. 1985 Dec;82(23):7989–7993. doi: 10.1073/pnas.82.23.7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock J. B., Ninfa A. J., Stock A. M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989 Dec;53(4):450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu-Cuot P., Carlier C., Poyart-Salmeron C., Courvalin P. A pair of mobilizable shuttle vectors conferring resistance to spectinomycin for molecular cloning in Escherichia coli and in gram-positive bacteria. Nucleic Acids Res. 1990 Jul 25;18(14):4296–4296. doi: 10.1093/nar/18.14.4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu-Cuot P., Carlier C., Poyart-Salmeron C., Courvalin P. An integrative vector exploiting the transposition properties of Tn1545 for insertional mutagenesis and cloning of genes from gram-positive bacteria. Gene. 1991 Sep 30;106(1):21–27. doi: 10.1016/0378-1119(91)90561-o. [DOI] [PubMed] [Google Scholar]
- Walsh C. T. Enzymes in the D-alanine branch of bacterial cell wall peptidoglycan assembly. J Biol Chem. 1989 Feb 15;264(5):2393–2396. [PubMed] [Google Scholar]
- Wurtzel E. T., Chou M. Y., Inouye M. Osmoregulation of gene expression. I. DNA sequence of the ompR gene of the ompB operon of Escherichia coli and characterization of its gene product. J Biol Chem. 1982 Nov 25;257(22):13685–13691. [PubMed] [Google Scholar]
- al-Obeid S., Collatz E., Gutmann L. Mechanism of resistance to vancomycin in Enterococcus faecium D366 and Enterococcus faecalis A256. Antimicrob Agents Chemother. 1990 Feb;34(2):252–256. doi: 10.1128/aac.34.2.252. [DOI] [PMC free article] [PubMed] [Google Scholar]