Abstract
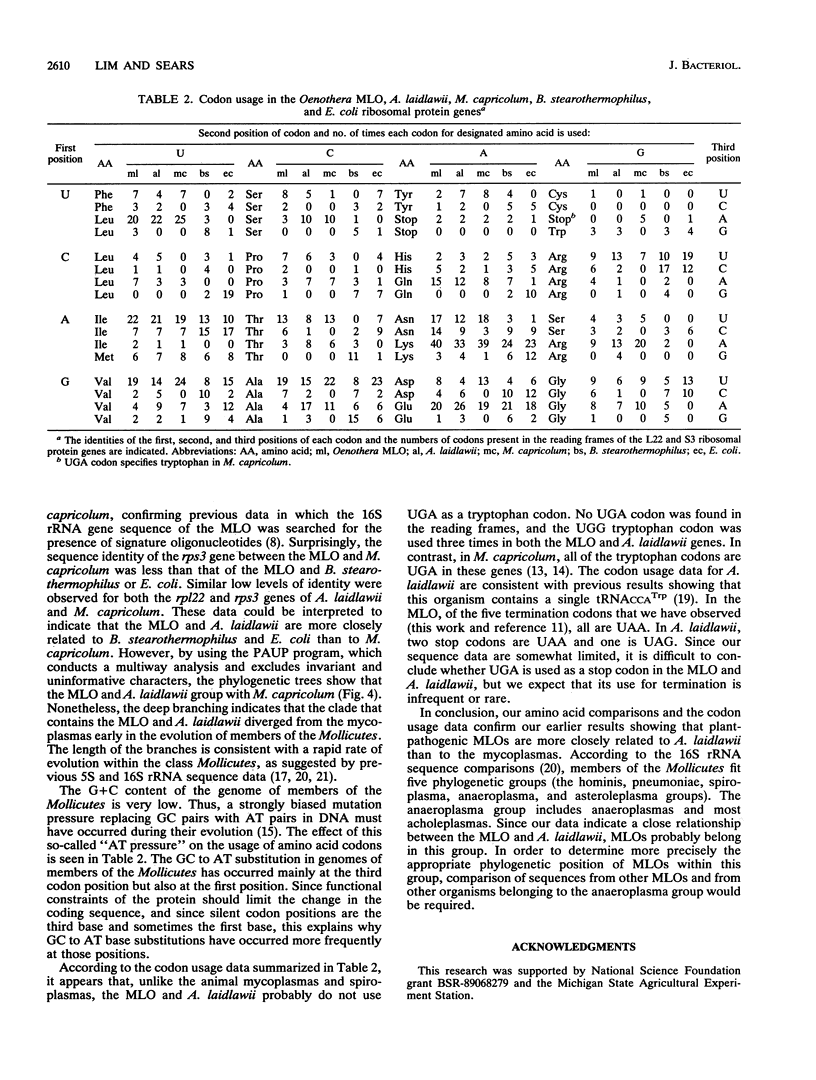
The families within the class Mollicutes are distinguished by their morphologies, nutritional requirements, and abilities to metabolize certain compounds. Biosystematic classification of the plant-pathogenic mycoplasmalike organisms (MLOs) has been difficult because these organisms have not been cultured in vitro, and hence their nutritional requirements have not been determined nor have physiological characterizations been possible. To investigate the evolutionary relationship of the MLOs to other members of the class Mollicutes, a segment of a ribosomal protein operon was cloned and sequenced from an aster yellows-type MLO which is pathogenic for members of the genus Oenothera and from Acholeplasma laidlawii. The deduced amino acid sequence data from the rpl22 and rps3 genes indicate that the MLOs are more closely related to A. laidlawii than to animal mycoplasmas, confirming previous results from 16S rRNA sequence comparisons. This conclusion is also supported by the finding that the UGA codon is not read as a tryptophan codon in the MLO and A. laidlawii, in contrast to its usage in Mycoplasma capricolum.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Huang M. N., High K. A. Efficient subcloning of DNA fragments amplified by crude oligonucleotides. Biotechniques. 1990 Dec;9(6):710–711. [PubMed] [Google Scholar]
- Inamine J. M., Ho K. C., Loechel S., Hu P. C. Evidence that UGA is read as a tryptophan codon rather than as a stop codon by Mycoplasma pneumoniae, Mycoplasma genitalium, and Mycoplasma gallisepticum. J Bacteriol. 1990 Jan;172(1):504–506. doi: 10.1128/jb.172.1.504-506.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol Rev. 1983 Mar;47(1):1–45. doi: 10.1128/mr.47.1.1-45.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krömer W. J., Hatakeyama T., Kimura M. Nucleotide sequences of Bacillus stearothermophilus ribosomal protein genes: part of the ribosomal S10 operon. Biol Chem Hoppe Seyler. 1990 Jul;371(7):631–636. doi: 10.1515/bchm3.1990.371.2.631. [DOI] [PubMed] [Google Scholar]
- Lim P. O., Sears B. B. 16S rRNA sequence indicates that plant-pathogenic mycoplasmalike organisms are evolutionarily distinct from animal mycoplasmas. J Bacteriol. 1989 Nov;171(11):5901–5906. doi: 10.1128/jb.171.11.5901-5906.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
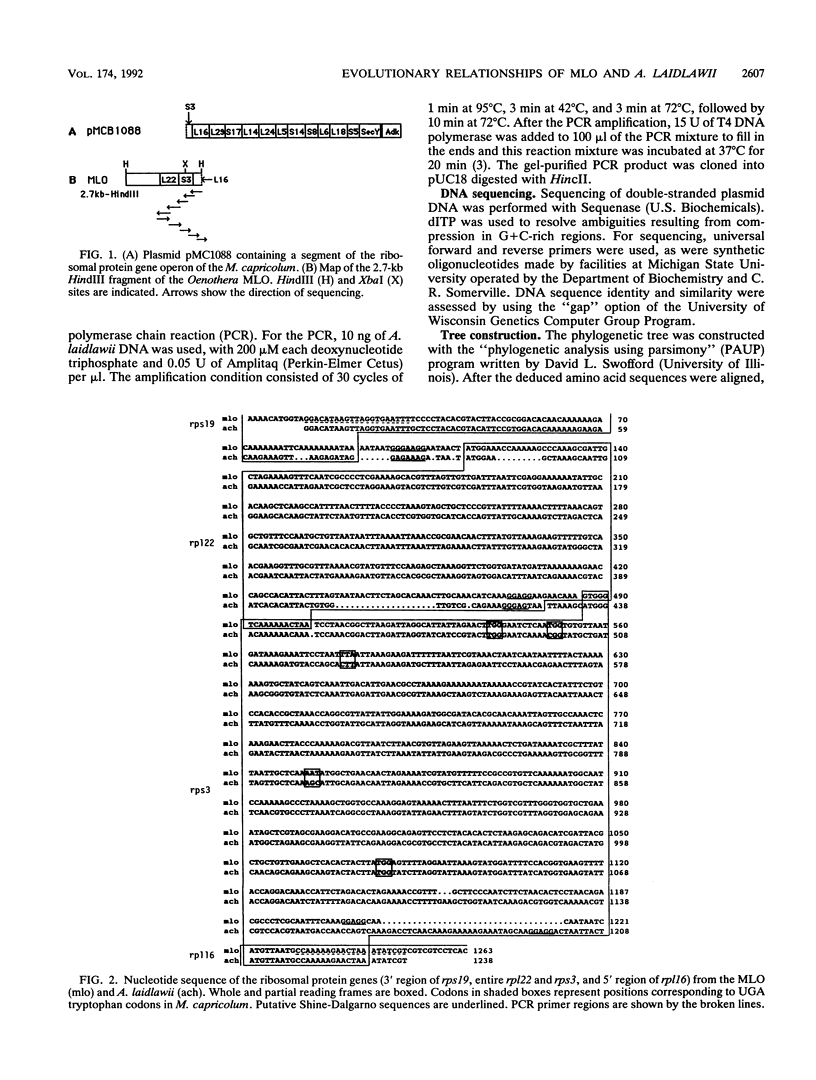
- Lim P. O., Sears B. B. DNA sequence of the ribosomal protein genes rp12 and rps19 from a plant-pathogenic mycoplasma-like organism. FEMS Microbiol Lett. 1991 Nov 1;68(1):71–73. doi: 10.1016/0378-1097(91)90397-s. [DOI] [PubMed] [Google Scholar]
- Lim P. O., Sears B. B. The genome size of a plant-pathogenic mycoplasmalike organism resembles those of animal mycoplasmas. J Bacteriol. 1991 Mar;173(6):2128–2130. doi: 10.1128/jb.173.6.2128-2130.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto A., Osawa S. The guanine and cytosine content of genomic DNA and bacterial evolution. Proc Natl Acad Sci U S A. 1987 Jan;84(1):166–169. doi: 10.1073/pnas.84.1.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkubo S., Muto A., Kawauchi Y., Yamao F., Osawa S. The ribosomal protein gene cluster of Mycoplasma capricolum. Mol Gen Genet. 1987 Dec;210(2):314–322. doi: 10.1007/BF00325700. [DOI] [PubMed] [Google Scholar]
- Osawa S., Jukes T. H. Codon reassignment (codon capture) in evolution. J Mol Evol. 1989 Apr;28(4):271–278. doi: 10.1007/BF02103422. [DOI] [PubMed] [Google Scholar]
- Razin S. Molecular biology and genetics of mycoplasmas (Mollicutes). Microbiol Rev. 1985 Dec;49(4):419–455. doi: 10.1128/mr.49.4.419-455.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers M. J., Simmons J., Walker R. T., Weisburg W. G., Woese C. R., Tanner R. S., Robinson I. M., Stahl D. A., Olsen G., Leach R. H. Construction of the mycoplasma evolutionary tree from 5S rRNA sequence data. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1160–1164. doi: 10.1073/pnas.82.4.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka R., Muto A., Osawa S. Nucleotide sequence of tryptophan tRNA gene in Acholeplasma laidlawii. Nucleic Acids Res. 1989 Jul 25;17(14):5842–5842. doi: 10.1093/nar/17.14.5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisburg W. G., Tully J. G., Rose D. L., Petzel J. P., Oyaizu H., Yang D., Mandelco L., Sechrest J., Lawrence T. G., Van Etten J. A phylogenetic analysis of the mycoplasmas: basis for their classification. J Bacteriol. 1989 Dec;171(12):6455–6467. doi: 10.1128/jb.171.12.6455-6467.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Maniloff J., Zablen L. B. Phylogenetic analysis of the mycoplasmas. Proc Natl Acad Sci U S A. 1980 Jan;77(1):494–498. doi: 10.1073/pnas.77.1.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamao F., Muto A., Kawauchi Y., Iwami M., Iwagami S., Azumi Y., Osawa S. UGA is read as tryptophan in Mycoplasma capricolum. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2306–2309. doi: 10.1073/pnas.82.8.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee T., Inouye M. Reexamination of the genome size of myxobacteria, including the use of a new method for genome size analysis. J Bacteriol. 1981 Mar;145(3):1257–1265. doi: 10.1128/jb.145.3.1257-1265.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurawski G., Zurawski S. M. Structure of the Escherichia coli S10 ribosomal protein operon. Nucleic Acids Res. 1985 Jun 25;13(12):4521–4526. doi: 10.1093/nar/13.12.4521. [DOI] [PMC free article] [PubMed] [Google Scholar]


