Abstract
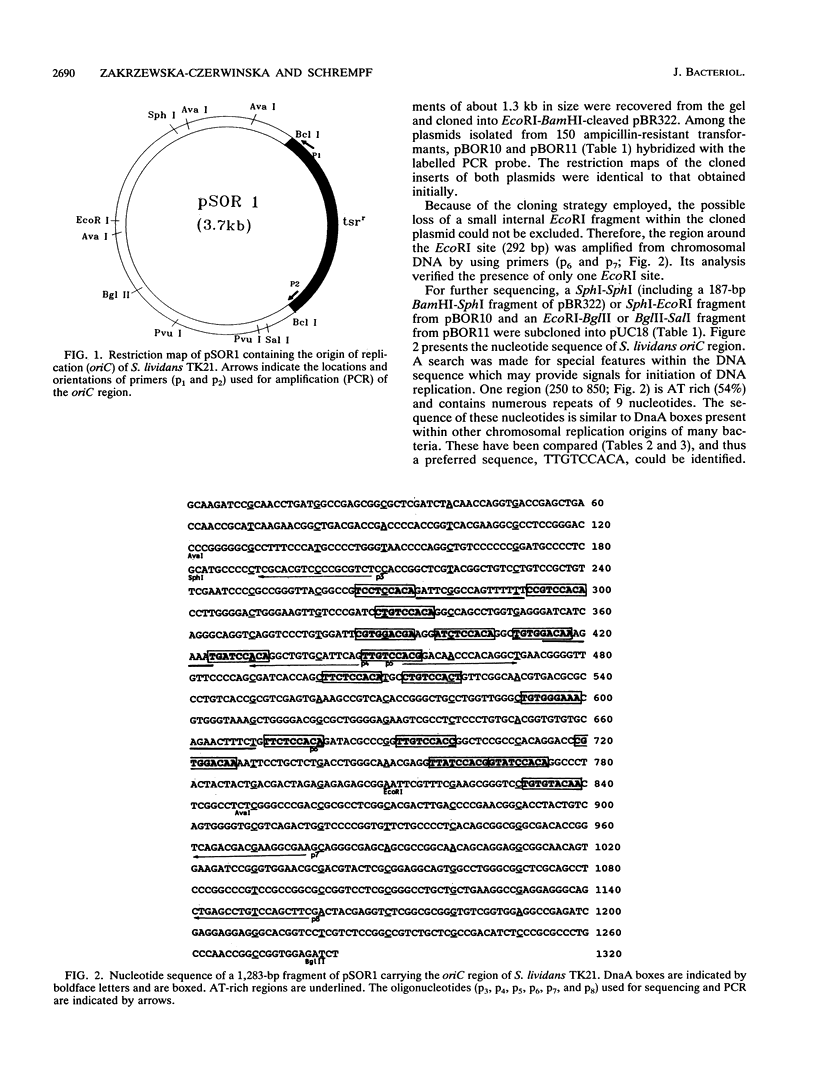
The chromosomal replication origin of the plasmidless derivative (TK21) from Streptomyces lividans 66 has been cloned as an autonomously replicating minichromosome (pSOR1) by using the thiostrepton resistance gene as a selectable marker. pSOR1 could be recovered as a closed circular plasmid which shows high segregational instability. pSOR1 was shown to replicate in Streptomyces coelicolor A3(2) and in S. lividans 66 and hybridized with DNA from several different Streptomyces strains. Physical mapping revealed that oriC is located on a 330-kb AseI fragment of the S. coelicolor A3(2) chromosome. DNA sequence analyses showed that the cloned chromosomal oriC region contains numerous DnaA boxes which are arranged in two clusters. The preferred sequence identified in the oriC region of Escherichia coli and several other bacteria is TTATCCACA. In contrast, in S. lividans, which has a high GC content, the preferred sequence for DnaA boxes appears to be TTGTCCACA.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnboim H. C. A rapid alkaline extraction method for the isolation of plasmid DNA. Methods Enzymol. 1983;100:243–255. doi: 10.1016/0076-6879(83)00059-2. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Bramhill D., Kornberg A. A model for initiation at origins of DNA replication. Cell. 1988 Sep 23;54(7):915–918. doi: 10.1016/0092-8674(88)90102-x. [DOI] [PubMed] [Google Scholar]
- Bramhill D., Kornberg A. Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell. 1988 Mar 11;52(5):743–755. doi: 10.1016/0092-8674(88)90412-6. [DOI] [PubMed] [Google Scholar]
- Fujita M. Q., Yoshikawa H., Ogasawara N. Structure of the dnaA region of Micrococcus luteus: conservation and variations among eubacteria. Gene. 1990 Sep 1;93(1):73–78. doi: 10.1016/0378-1119(90)90138-h. [DOI] [PubMed] [Google Scholar]
- Ogasawara N., Moriya S., Yoshikawa H. Initiation of chromosome replication: structure and function of oriC and DnaA protein in eubacteria. Res Microbiol. 1991 Sep-Oct;142(7-8):851–859. doi: 10.1016/0923-2508(91)90065-i. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer C., Messer W. DnaA protein/DNA interaction. Modulation of the recognition sequence. Mol Gen Genet. 1991 Apr;226(1-2):34–40. doi: 10.1007/BF00273584. [DOI] [PubMed] [Google Scholar]
- Schwartz D. C., Cantor C. R. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984 May;37(1):67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- Thompson C. J., Kieser T., Ward J. M., Hopwood D. A. Physical analysis of antibiotic-resistance genes from Streptomyces and their use in vector construction. Gene. 1982 Nov;20(1):51–62. doi: 10.1016/0378-1119(82)90086-5. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yasuda S., Hirota Y. Cloning and mapping of the replication origin of Escherichia coli. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5458–5462. doi: 10.1073/pnas.74.12.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee T. W., Smith D. W. Pseudomonas chromosomal replication origins: a bacterial class distinct from Escherichia coli-type origins. Proc Natl Acad Sci U S A. 1990 Feb;87(4):1278–1282. doi: 10.1073/pnas.87.4.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakrzewska-Czerwińska J., Gaszewska A., Mordarski M. pS10147-2, a 3.7 kb multicopy plasmid isolated from Streptomyces coelicolor. FEMS Microbiol Lett. 1990 Sep 15;59(3):271–275. doi: 10.1016/0378-1097(90)90232-f. [DOI] [PubMed] [Google Scholar]
- Zyskind J. W., Cleary J. M., Brusilow W. S., Harding N. E., Smith D. W. Chromosomal replication origin from the marine bacterium Vibrio harveyi functions in Escherichia coli: oriC consensus sequence. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1164–1168. doi: 10.1073/pnas.80.5.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zyskind J. W., Deen L. T., Smith D. W. Isolation and mapping of plasmids containing the Salmonella typhimurium origin of DNA replication. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3097–3101. doi: 10.1073/pnas.76.7.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]