Abstract
The large docking protein IRS-1 is a major substrate for the insulin receptor and other tyrosine kinases. It plays a key role in eliciting many of insulin’s actions, including binding and activation of phosphatidylinositol (PI) 3-kinase and the subsequent increase in glucose transport. Gene disruption of IRS-1 in mice is associated with an impaired insulin-stimulated glucose disposal in vivo and glucose transport in vitro, but the survival of the animals and residual insulin sensitivity is dependent on the presence of the alternative docking protein IRS-2. We examined the expression and function of IRS-1 and IRS-2 in adipocytes from healthy and diabetic individuals. Cells from subjects with non-insulin-dependent diabetes mellitus (NIDDM), but not with insulin-dependent diabetes mellitus, had an impaired insulin effect and a marked reduction (70 ± 6%) in the expression of IRS-1 protein, whereas IRS-2 was unchanged. In normal cells, IRS-1 was the main docking protein for the binding and activation of insulin-stimulated PI 3-kinase; IRS-2 was also functional but required a higher insulin concentration for a similar binding and activation of PI 3-kinase. In contrast in NIDDM cells with a low IRS-1 content, IRS-2 became the main docking protein. These findings may provide important reasons for the insulin resistance in NIDDM.
Insulin resistance in various target tissues and an insufficient compensatory increase in insulin release by the β cells are the main causes of non-insulin-dependent diabetes mellitus (NIDDM) (1, 2).
Insulin plays a key role for the regulation of metabolism in many mammalian cells–principally, liver, muscle, and adipose cells (3, 4). The ability of insulin to increase glucose transport into muscle and fat cells is mediated by the translocation of a specific glucose transporter, GLUT4, from intracellular vesicles to the cell surface (5–7).
The initial mechanism of insulin action involves its binding to specific cell surface receptors leading to the autophosphorylation and activation of an intrinsic tyrosine kinase associated with the β-receptor subunit. IRS proteins (IRS-1 and IRS-2) are substrates for the insulin receptor and other tyrosine kinases associated with the receptors of growth factors and cytokines (1, 8–12). IRS proteins act as an interface between activated receptors and signaling proteins with Src homology 2 (SH2) domains. After insulin stimulation, IRS-1 associates with several proteins including phosphatidylinositol (PI) 3kinase, Syp, Nck, Grb2, and Fyn (13–16). PI 3-kinase is a heterodimeric enzyme consisting of an 85-kDa regulatory subunit with SH2 domains capable of binding to tyrosine-phosphorylated IRS-1 and a 110-kDa catalytic subunit that phosphorylates the inositol ring of PI and its phosphorylated derivatives (17, 18). PI 3-kinase has been implicated as one of the key signal transducers in insulin-stimulated glucose uptake and GLUT4 translocation (19–23). Little is known about the roles of IRS proteins in the regulation of PI 3-kinase binding and activation in normal physiology or in insulin-resistant states in human subjects.
In this study, we examined the expression and function of IRS-1 and IRS-2 in adipocytes from healthy and diabetic individuals. IRS-1 was the main docking protein for PI 3kinase in adipocytes from healthy subjects. However, in adipocytes from NIDDM subjects, IRS-1 protein content was markedly reduced and IRS-2 became the main docking protein. Binding of PI 3-kinase to IRS-2 required a higher insulin concentration than that needed for a similar binding to IRS-1 and this may be a key factor for the insulin resistance in NIDDM.
MATERIALS AND METHODS
Glucose Transport in Human Adipocytes.
Specimens of human subcutaneous adipose tissue were obtained from the abdominal region of nondiabetic [n = 14, 4 men and 10 women; age, 44 ± 3 years; body mass index (BMI), 30 ± 2 kg/m2], NIDDM (n = 12, 8 men and 4 women; age, 63 ± 2 years; BMI, 29.9 ± 0.9 kg/m2; HbA1c, 7.1 ± 1.1%; reference value, 5.3%), or insulin-dependent diabetes mellitus (IDDM; n = 8, 6 men and 2 women; age, 41 ± 5 years; BMI, 27.8 ± 1.2 kg/m2; HbA1c, 7.4 ± 0.6%) subjects. The biopsies were placed in Medium 199 at 37°C containing 25 mM Hepes, 4% BSA, and 5.5 mM glucose. The study was approved by the Ethical Committee of the Goteborg University.
Adipose cells were prepared as described (24). Briefly, the adipocytes were isolated by digesting about 0.6 g of tissue for 50 min at 37°C in Medium 199 containing 25 mM Hepes, 4% BSA, 5.5 mM glucose, and collagenase (Sigma) at 0.8 mg/ml in a shaking water bath. Cells were then incubated for 15 min in the absence or presence of the indicated concentrations of human insulin (Novo-Nordisk, Copenhagen), 0.1 μM N6-(2-phenylisopropyl)adenosine, and adenosine deaminase (1 unit/ml). Glucose transport activity was assayed for 1 h with 0.86 μM [U-14C]glucose (Amersham) as described (25). The cells were separated from the incubation medium by centrifugation through silicone oil and the radioactivity associated with the cells was measured by scintillation counting.
Immunoprecipitations and Immunoblotting.
Isolated human adipocytes were distributed into plastic vials (12–15% cell suspension) in a final incubation volume of 400 μl. Cells were preincubated with or without 6.9 nM insulin for 10 min, immediately separated by centrifugation through silicone oil, and lysed in 0.4 ml of lysis buffer containing 25 mM Tris·HCl (pH 7.4), 0.5 mM EGTA, 25 mM NaCl, 1% Nonidet P-40, 1 mM Na3VO4, 10 mM NaF, 0.2 mM leupeptin, 1 mM benzamidine, and 0.1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride and rocked for 40 min at 4°C. Detergent-insoluble material was sedimented by centrifugation at 12,000 × g for 10 min at 4°C. Cell lysate proteins (50 μg of protein) were separated by SDS/PAGE or 100 μg of protein was immunoprecipitated for 2 h with anti-IRS-1 C-terminal (4 μg/ml; Upstate Biotechnology, Lake Placid, NY) or anti-IRS-2 (3 μl/ml) antibodies. Antibodies against IRS-2 were prepared in rabbits by using an IRS-2-specific peptide composed of amino acids 1310–1322 (LSHHLKEATVVKE). Immune complexes were collected with protein A-Sepharose, washed, solubilized in Laemmli sample buffer, and separated by SDS/PAGE on 7.5% gels. Proteins were transferred from the gel to nitrocellulose sheets and blocked in 5% milk. The blots were probed with various primary antibodies as follows: anti-IRS-1 C-terminal, anti-p85 (whole antiserum), anti-SHPTP2/syp, anti-Grb2 and 4G10 anti-phosphotyrosine antibodies (Upstate Biotechnology); anti-insulin receptor (Transduction Laboratories, Lexington, KY), anti-IRS-1 (N-terminal), or anti-p110 (Santa Cruz Biotechnology) according to the recommendations of the manufacturer or anti-IRS-2 (1:500 dilution). The proteins were detected by enhanced chemiluminescence and horseradish peroxidase-labeled second antibodies (Amersham). The intensity of the bands was quantitated with a laser densitometer (Molecular Dynamics). Quantification was also verified with 125I-labeled protein A. Control experiments showed that the antibodies to IRS-1 and IRS-2 had a similar efficiency (≈90%) to deplete their respective antigens.
PI 3-Kinase Activity.
PI 3-kinase assay was performed directly on the immunoprecipitates as described (25, 26). Briefly, 6 μl of a mixture of PI (10 μg/sample) and phosphatidylserine (2.5 μg/sample) were added to the beads and the reaction was started by the addition of 30 μl of a reaction mixture consisting of 40 mM Hepes (pH 7.5), 20 mM MgCl2, and 50 μM [γ-32P]ATP (0.2 μCi/μl; 1 Ci = 37 GBq). After 15 min at 30°C, the reaction was stopped by the addition of 40 μl of 4 M HCl and 160 μl of CHCl3/CH3OH (1:1). The organic phase was extracted and applied to a silica gel thin layer chromatography plate precoated with 1% potassium oxalate (Analtech). The chromatography plates were developed in CHCl3/CH3OH/H2O/NH4OH (60:47:11.3:2), dried, and visualized by autoradiography. The radioactivity was quantitated with a PhosphorImager (Molecular Dynamics).
RESULTS AND DISCUSSION
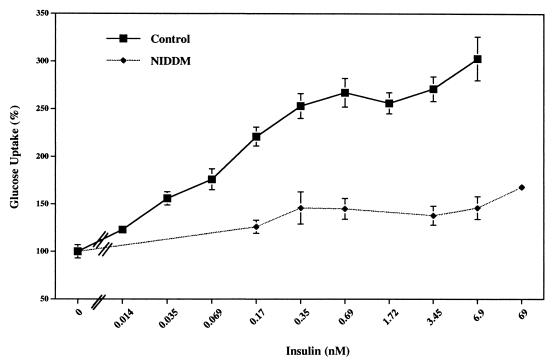
We investigated the expression and function of the IRS proteins in adipocytes isolated from healthy subjects and diabetic patients, both NIDDM and IDDM. First, we measured the glucose transport activity in response to different concentrations of insulin (Fig. 1). The half-maximal response in healthy subjects was at 0.13 nM insulin and the maximal response (≈3 times over basal) was at 0.35 nM insulin. In contrast, in cells from NIDDM patients, both the concentration required to significantly increase glucose transport and the maximal response were significally impaired with only 50% increase with respect to basal transport at maximal concentrations of insulin (Fig. 1).
Figure 1.
Effect of various concentrations of insulin on glucose transport in adipocytes from healthy subjects and NIDDM subjects. Values represent the mean ± SEM of six experiments with triplicate determinations. The value at 100% is the basal uptake in the absence of insulin.
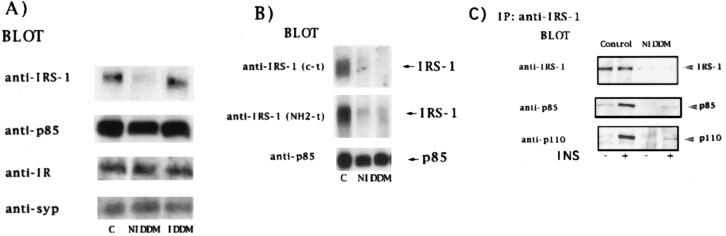
To determine whether the impairment in insulin action in adipocytes from NIDDM subjects was associated with an abnormality in proteins involved in insulin signaling, immunoblotting was performed with antibodies against several proteins (Fig. 2A). IRS-1 was reduced by 50–90% (average, 70 ± 6%) in adipocytes from NIDDM patients compared with those from nondiabetic subjects. This was confirmed by using two different N- and C-terminal antibodies (Fig. 2B). However, the insulin receptor, the p85 subunit of PI 3-kinase, and the phosphotyrosine phosphatase SHPTP2/Syp (Fig. 2A) were unchanged. Similarly, the IRS-1 protein content was not reduced in adipocytes from IDDM subjects. As expected, in IRS-1 immunoprecipitates from NIDDM cell lysates, proteins corresponding to IRS-1 or PI 3-kinase (p85 and p110 subunits) in response to insulin could hardly be detected (Fig. 2C).
Figure 2.
IRS-1 protein content in adipocytes from healthy subjects and IDDM and NIDDM subjects. (A and B) Cell lysates from adipocytes of healthy control, IDDM, or NIDDM subjects (50 μg of protein) were separated by SDS/PAGE (10% polyacrylamide gels) and immunoblotted with the indicated antibodies. Each lane represents the pooled lysates of two subjects, and each blot is representative of at least three experiments. (C) Cell lysates from adipocytes preincubated in the presence or absence of insulin (INS) were immunoprecipitated with antibodies against IRS-1 (C-terminal), separated by SDS/PAGE on 7.5% gels, and immunoblotted with the indicated antibodies.
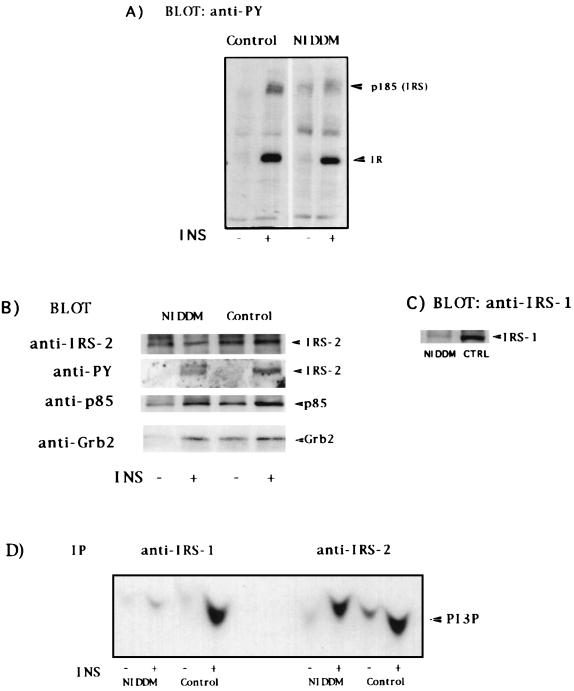
Despite the big reduction of IRS-1, immunoblots with anti-phosphotyrosine antibodies in whole cell lysates from basal or insulin-stimulated NIDDM adipocytes showed a tyrosine-phosphorylated protein at ≈185 kDa, suggesting the presence of another protein at that position (Fig. 3A). The likely candidate was the recently identified docking protein IRS-2 (27). In immunoprecipitates with polyclonal IRS-2 antibodies, IRS-2 was present to a similar extent in adipocytes from both NIDDM and healthy subjects (Fig. 3B); IRS-2 was also tyrosine-phosphorylated in an insulin-dependent fashion. It was clear that both PI 3-kinase and Grb2 became associated with IRS-2 in response to insulin (Fig. 3B) as reported (28, 29). Supernatants from anti-IRS-2 immunoprecipitations analyzed by immunoblotting again showed that IRS-1 was barely detectable in NIDDM cells (Fig. 3C).
Figure 3.
Expression and function of IRS-2 in adipocytes from healthy control and NIDDM subjects. (A) Adipocytes from control or NIDDM subjects were incubated without or with 6.9 nM insulin (INS) for 10 min. Cell lysates were loaded by equal amount of proteins, separated by SDS/PAGE (10% gels), and immunoblotted with anti-phosphotyrosine (PY) antibodies. (B) Cell lysates were immunoprecipitated with anti-IRS-2 antibodies and immunoblotted with anti-IRS-2, anti-PY, anti-p85, and anti-Grb2 as indicated. (C) Supernatants from anti-IRS-2 immunoprecipitates were separated by SDS/PAGE (7.5% gels) and immunoblotted with anti-IRS-1 C-terminal antibodies. (D) PI 3-kinase activity was assayed on immunoprecipitates (IP) produced by the indicated antibodies on lysates from untreated or insulin-stimulated adipocytes from control or NIDDM subjects.
The PI 3-kinase activity recovered in IRS-1 and IRS-2 immunoprecipitates was markedly stimulated in normal adipocytes by insulin (Fig. 3D). Again, little or no PI 3-kinase activity was recovered in IRS-1 immunoprecipitates from NIDDM adipocytes. In these cells, PI 3-kinase activity was virtually exclusively recovered in the anti-IRS-2 immunoprecipitates (Fig. 3D).
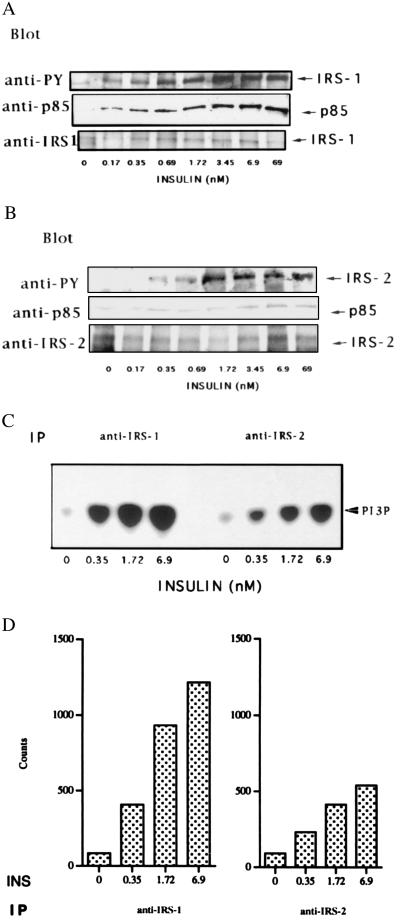
We next examined the concentration-dependent ability of insulin to increase the tyrosine phosphorylation of IRS-1 and IRS-2 and the subsequent binding of PI 3-kinase in adipocytes from healthy subjects. Both IRS-1 and IRS-2 became tyrosine-phosphorylated in the presence of insulin but quantification of the immunoblots showed that the effect of a maximal insulin concentration was ≈50% greater for IRS-1 than for IRS-2. In agreement with this finding, immunoprecipitates with anti-IRS-1 or anti-IRS-2 antibodies showed that at all insulin concentrations IRS-1 was the main docking protein for PI 3-kinase (Fig. 4A). However, IRS-2 could also bind PI 3-kinase (Fig. 4B) but this was, for a given insulin concentration, less (≈40%) than that bound to IRS-1. This provides evidence for differences in insulin-stimulated phosphorylation of IRS-1 and IRS-2 that may be related to the different interaction between IRS-2 and the insulin receptor (30, 31). This was further verified when we measured the PI 3-kinase lipid kinase activity. At all insulin concentrations, IRS-1 accounts for most (≈70%) of the IRS-associated PI 3-kinase activity in response to insulin in adipocytes from normal subjects (Fig. 4C). In contrast, IRS-2 was the main source of PI 3-kinase activity in adipocytes from NIDDM subjects (Fig. 3D).
Figure 4.
IRS-1 is the main docking protein for PI 3-kinase in human adipocytes. Adipocytes from healthy control subjects were incubated for 10 min with different concentrations of insulin. Cell lysates were immunoprecipitated with anti-IRS-1 C-terminal antibodies (A) or anti-IRS-2 antibodies (B) separated on 7.5% SDS-PAGE and immunoblotted with the indicated antibodies. (C) PI 3-kinase activity was assayed on immunoprecipitates produced by the indicated antibodies as described in Fig. 3. (D) Quantification of the PI 3-kinase activity using a PhosphorImager (Molecular Dynamics).
In this study we demonstrate that IRS-1 protein expression is markedly reduced in adipocytes from NIDDM subjects in comparison with both healthy subjects and individuals with IDDM. The 50% or greater reduction in IRS-1 content was consistently seen in the 12 NIDDM subjects studied. Subsequently, we have examined more than 20 NIDDM patients and only seen a smaller reduction (≈30%) in two subjects. This reduction cannot be accounted for by obesity since the control and NIDDM subjects had a similar BMI or by hyperinsulinemia/hyperglycemia since the IDDM were unaffected. In contrast, IRS-2 levels were unchanged in NIDDM cells. Skeletal muscle from morbidly obese individuals has also been shown, at an average, to have a moderate reduction in IRS-1 protein content (32) but it is not clear whether this reduction was confined to subjects with an impaired glucose tolerance.
IRS-1 seems to be the main docking protein for PI 3-kinase and the associated increase in glucose uptake in normal human adipocytes, as also demonstrated in other cells (33–35). However, similar to the IRS-1-deficient mice (28, 36, 37), IRS-2 seems to be able to replace IRS-1 as the main docking protein for binding and activation of PI 3-kinase. However, in human adipocytes, IRS-2 requires a higher insulin concentration than IRS-1 for a similar binding and activation of PI 3-kinase. This is in parallel to the ability of insulin to increase total tyrosine phosphorylation, which also was reduced in IRS-2. These findings may provide important reasons for the insulin resistance in NIDDM.
The basic mechanism(s) for the reduction in IRS-1 expression in NIDDM is currently unclear. However, preliminary evidence indicates that IRS-1 mRNA levels are reduced in adipocytes from NIDDM subjects. Whether this is sufficient to account for the low IRS-1 content or whether there also is an increased protein degradation is unclear. We are currently exploring potential mechanisms and examining IRS-1 content in human adipocytes from different nondiabetic but insulin-resistant states.
Acknowledgments
We thank Drs. C. Londos and S. Cushman for helpful comments and discussion. U.S. was a Scholar in Residence at the National Institutes of Health. This study was supported by grants from the Swedish Medical Research Council, the Swedish Diabetes Foundation, the Novo-Nordisk Foundation, the Tore Nilsons Foundation, and the IngaBritt and Arne Lundberg Foundation.
ABBREVIATIONS
- BMI
body mass index
- IRS
insulin receptor substrate
- PI
phosphatidylinositol
- NIDDM
non-insulin-dependent diabetes mellitus
- IDDM
insulin-dependent diabetes mellitus
References
- 1.Eriksson J, Franssila-Kallunki A, Ekstrand A, Saloranta C, Widen E, Schalin C, Groop L. N Engl J Med. 1989;321:337–343. doi: 10.1056/NEJM198908103210601. [DOI] [PubMed] [Google Scholar]
- 2.Martin B C, Warram J H, Krolewski A S, Bergman R N, Soeldner J S, Kahn C R. Lancet. 1992;340:925–929. doi: 10.1016/0140-6736(92)92814-v. [DOI] [PubMed] [Google Scholar]
- 3.White M F, Kahn C R. J Biol Chem. 1994;269:1–4. [PubMed] [Google Scholar]
- 4.Kahn C R. Diabetes. 1994;43:1066–1084. doi: 10.2337/diab.43.8.1066. [DOI] [PubMed] [Google Scholar]
- 5.Cushman S W, Wardzala L J. J Biol Chem. 1980;255:4758–4762. [PubMed] [Google Scholar]
- 6.Suzuki K, Kono T. Proc Natl Acad Sci USA. 1980;77:2542–2545. doi: 10.1073/pnas.77.5.2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.James D E, Strube M, Mueckler M. Nature (London) 1989;338:83–87. doi: 10.1038/338083a0. [DOI] [PubMed] [Google Scholar]
- 8.White M F, Maron R, Kahn C R. Nature (London) 1985;318:183–186. doi: 10.1038/318183a0. [DOI] [PubMed] [Google Scholar]
- 9.Sun X-J, Rothenberg P, Kahn C R, Backer J M, Araki E, Wilden P A, Cahill A, Goldstein B J, White M F. Nature (London) 1991;352:73–77. doi: 10.1038/352073a0. [DOI] [PubMed] [Google Scholar]
- 10.Wang L-M, Myers M G, Jr, Sun X, Aaronson S A, White M F, Pierce J H. Science. 1993;261:1591–1594. doi: 10.1126/science.8372354. [DOI] [PubMed] [Google Scholar]
- 11.Myers M G, Sun X J, White M F. Trends Biochem Sci. 1994;19:289–294. doi: 10.1016/0968-0004(94)90007-8. [DOI] [PubMed] [Google Scholar]
- 12.Sun X J, Crimmins D L, Myers M G, Miralpeix M, White M F. Mol Cell Biol. 1993;13:7418–7428. doi: 10.1128/mcb.13.12.7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Myers M G, Jr, Backer J M, Suin X J, Shoelson S E, Hu P, Schlessinger J, Yoakim M, Schaffhausen B, White M F. Proc Natl Acad Sci USA. 1992;89:10350–10354. doi: 10.1073/pnas.89.21.10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Myers M G, Jr, Wang L M, Sun X, Zhang Y, Yenush L P, Schlessinger J, Pierce J H, White M F. Mol Cell Biol. 1994;14:3577–3587. doi: 10.1128/mcb.14.6.3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noguchi T, Matozaki T, Horita K, Fujioka Y, Kasuga M. Mol Cell Biol. 1994;14:6674–6682. doi: 10.1128/mcb.14.10.6674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuhne M R, Pawson T, Lienhard G E, Feng G-S. J Biol Chem. 1993;268:11479–11481. [PubMed] [Google Scholar]
- 17.Kapeller R, Cantley L C. BioEssays. 1994;16:565–576. doi: 10.1002/bies.950160810. [DOI] [PubMed] [Google Scholar]
- 18.Whitman M, Downes C P, Keeler M, Kellert T, Cantley L. Nature (London) 1988;332:644–646. doi: 10.1038/332644a0. [DOI] [PubMed] [Google Scholar]
- 19.Kanai F, Ito K, Todaka M, Hayashi H, Kamohara S, Ishii K, Okada T, Hazeki O, Ui M, Ebina Y. Biochem Biophys Res Commun. 1994;195:762–768. doi: 10.1006/bbrc.1993.2111. [DOI] [PubMed] [Google Scholar]
- 20.Okada T, Kawano Y, Sakakibara T, Hazeki O, Ui M. J Biol Chem. 1994;269:3568–3573. [PubMed] [Google Scholar]
- 21.Clark J F, Young P W, Yonezawa K, Fasuga M, Holman G D. Biochem J. 1994;300:631–635. doi: 10.1042/bj3000631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheatham B, Vlahos C J, Cheatham L, Wang L, Blenis J, Kahn C R. Mol Cell Biol. 1994;14:4902–4911. doi: 10.1128/mcb.14.7.4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hara K, Yonezawa K, Sakaue H, Ando A, Kotani K, Kitamura T, Kitamura Y, Ueda H, Stephens L, Jackson T R, Hawkins P T, Dhand R, Clark A E, Holman G D, Waterfield M D, Kasuga M. Proc Natl Acad Sci USA. 1994;91:7415–7419. doi: 10.1073/pnas.91.16.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith U, Sjostrom L, Bjorntorp P. J Lipid Res. 1972;13:822–827. [PubMed] [Google Scholar]
- 25.Rondinone C M, Smith U. J Biol Chem. 1996;271:18148–18153. doi: 10.1074/jbc.271.30.18148. [DOI] [PubMed] [Google Scholar]
- 26.Auger K R, Serunian L A, Soltoff S P, Libby P, Cantley L C. Cell. 1989;57:167–175. doi: 10.1016/0092-8674(89)90182-7. [DOI] [PubMed] [Google Scholar]
- 27.Sun X-J, Rothenberg P, Kahn C R, Backer J M, Araki E, Wilden P A, Cahill D A, Goldstein B J, White M F. Nature (London) 1991;352:73–77. doi: 10.1038/352073a0. [DOI] [PubMed] [Google Scholar]
- 28.Patti M E, Sun X J, Bruening J C, Araki E, Lipes M A, White M F, Kahn C R. J Biol Chem. 1995;270:24670–24673. doi: 10.1074/jbc.270.42.24670. [DOI] [PubMed] [Google Scholar]
- 29.Tobe K, Tamemoto H, Yamauchi T, Aizawa S, Yazaki Y, Kadowaki T. J Biol Chem. 1995;270:5698–5701. doi: 10.1074/jbc.270.11.5698. [DOI] [PubMed] [Google Scholar]
- 30.He W, Craparo A, Zhu Y, O’Neill T J, Wang L-M, Pierce J H, Gustafson T A. J Biol Chem. 1996;271:11641–11645. doi: 10.1074/jbc.271.20.11641. [DOI] [PubMed] [Google Scholar]
- 31.Sawka-Verrhelle D J, Tartare-Deckert S, White M F, Van Obberghen E. J Biol Chem. 1996;271:5980–5983. doi: 10.1074/jbc.271.11.5980. [DOI] [PubMed] [Google Scholar]
- 32.Goodyear L J, Giorgino F, Sherman L A, Carey J, Smith R J, Dohm G L. J Clin Invest. 1995;95:2195–2204. doi: 10.1172/JCI117909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quon M J, Butte A J, Zarnowski M J, Sesti G, Cushman S W, Taylor S I. J Biol Chem. 1994;269:27920–27924. [PubMed] [Google Scholar]
- 34.Hara K, Yonezawa K, Sakaue H, Ando A, Kotani K, Kitamura T, Kitamura Y, Ueda H, Stephens L, Jackson T R, Hawkins P T, Dhand R, Clark A E, Holman G D, Waterfield M D, Kasuga M. Proc Natl Acad Sci USA. 1994;91:7415–7419. doi: 10.1073/pnas.91.16.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quon M J, Chen H, Ing B L, Liu M L, Zarnowski M J, Yonezawa K, Kasuga M, Cushman S W, Taylor S I. Mol Cell Biol. 1995;15:5403–5411. doi: 10.1128/mcb.15.10.5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tamemoto H, Kadowaki T, Tobe K, Yagi T, Sakura H, Hayakawa T, Terauchi Y, Ueki K, Kaburagi Y, Satoh S, Sekihara H, Yoshioka S, Horikoshi H, Furuta Y, Ikawa Y, Kasuga M, Yazaki Y, Aizawa S. Nature (London) 1994;372:182–186. doi: 10.1038/372182a0. [DOI] [PubMed] [Google Scholar]
- 37.Araki E, Lipes M A, Pattii M-E, Bruening J C, Haag B J, III, Johnson R S, Kahn C R. Nature (London) 1994;372:186–190. doi: 10.1038/372186a0. [DOI] [PubMed] [Google Scholar]