Abstract
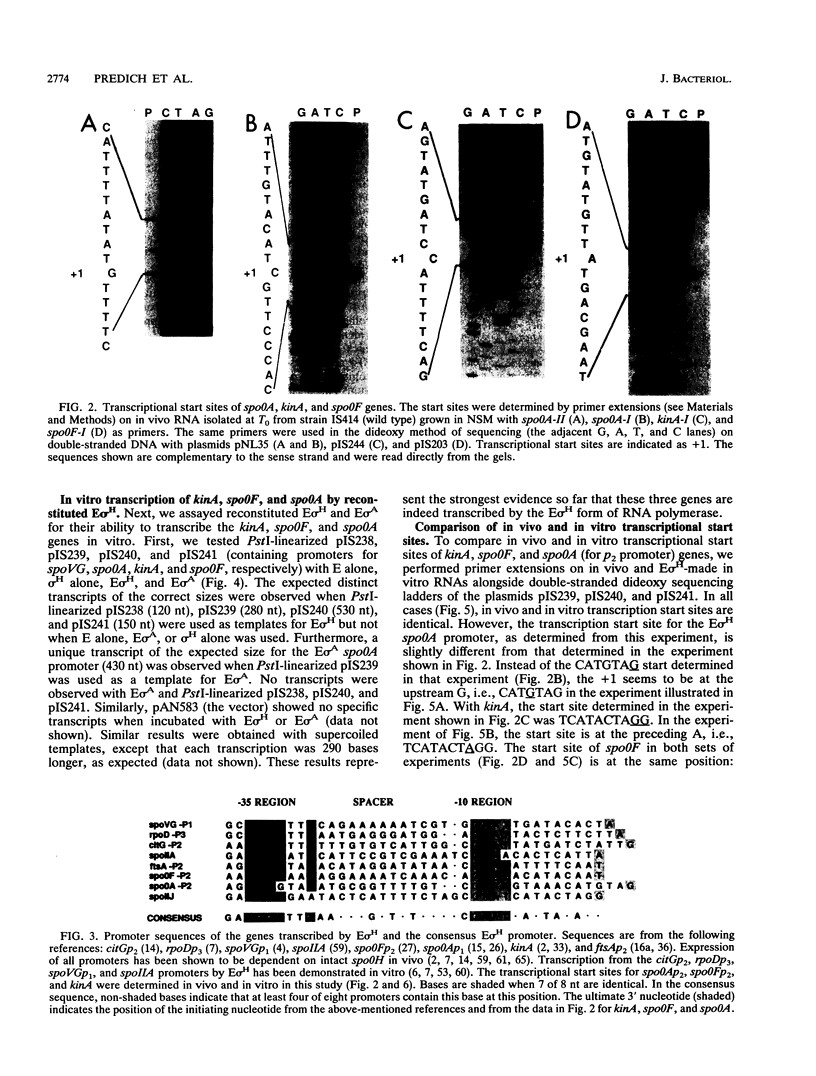
The Bacillus subtilis genes kinA (spoIIJ), spo0F, and spo0A encode components of the sporulation signal transduction pathway. Recent work has suggested that these genes are transcribed by a minor form of RNA polymerase, E sigma H (sigma H is the product of spo0H, another early sporulation gene). We directly tested this hypothesis by performing in vitro transcription assays with reconstituted E sigma H and a set of plasmids containing the kinA, spo0F, and spo0A promoter regions. We were able to obtain distinct transcripts of the expected sizes with all three genes by using linearized or supercoiled templates. Furthermore, primer extension experiments indicate that the transcription start sites for the three genes in vitro and in vivo are the same. In addition, we measured steady-state levels of kinA, spo0F, and spo0A mRNAs during growth in sporulation medium; all of them were increased at or near the beginning of the stationary phase.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aiba H., Mizuno T., Mizushima S. Transfer of phosphoryl group between two regulatory proteins involved in osmoregulatory expression of the ompF and ompC genes in Escherichia coli. J Biol Chem. 1989 May 25;264(15):8563–8567. [PubMed] [Google Scholar]
- Antoniewski C., Savelli B., Stragier P. The spoIIJ gene, which regulates early developmental steps in Bacillus subtilis, belongs to a class of environmentally responsive genes. J Bacteriol. 1990 Jan;172(1):86–93. doi: 10.1128/jb.172.1.86-93.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai U., Lewandoski M., Dubnau E., Smith I. Temporal regulation of the Bacillus subtilis early sporulation gene spo0F. J Bacteriol. 1990 Sep;172(9):5432–5439. doi: 10.1128/jb.172.9.5432-5439.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banner C. D., Moran C. P., Jr, Losick R. Deletion analysis of a complex promoter for a developmentally regulated gene from Bacillus subtilis. J Mol Biol. 1983 Aug 5;168(2):351–365. doi: 10.1016/s0022-2836(83)80023-0. [DOI] [PubMed] [Google Scholar]
- Burbulys D., Trach K. A., Hoch J. A. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell. 1991 Feb 8;64(3):545–552. doi: 10.1016/0092-8674(91)90238-t. [DOI] [PubMed] [Google Scholar]
- Carter H. L., 3rd, Moran C. P., Jr New RNA polymerase sigma factor under spo0 control in Bacillus subtilis. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9438–9442. doi: 10.1073/pnas.83.24.9438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter H. L., 3rd, Wang L. F., Doi R. H., Moran C. P., Jr rpoD operon promoter used by sigma H-RNA polymerase in Bacillus subtilis. J Bacteriol. 1988 Apr;170(4):1617–1621. doi: 10.1128/jb.170.4.1617-1621.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang B. Y., Doi R. H. Overproduction, purification, and characterization of Bacillus subtilis RNA polymerase sigma A factor. J Bacteriol. 1990 Jun;172(6):3257–3263. doi: 10.1128/jb.172.6.3257-3263.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibazakura T., Kawamura F., Takahashi H. Differential regulation of spo0A transcription in Bacillus subtilis: glucose represses promoter switching at the initiation of sporulation. J Bacteriol. 1991 Apr;173(8):2625–2632. doi: 10.1128/jb.173.8.2625-2632.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels D., Zuber P., Losick R. Two amino acids in an RNA polymerase sigma factor involved in the recognition of adjacent base pairs in the -10 region of a cognate promoter. Proc Natl Acad Sci U S A. 1990 Oct;87(20):8075–8079. doi: 10.1073/pnas.87.20.8075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison B. L., Leighton T., Rabinowitz J. C. Purification of Bacillus subtilis RNA polymerase with heparin-agarose. In vitro transcription of phi 29 DNA. J Biol Chem. 1979 Sep 25;254(18):9220–9226. [PubMed] [Google Scholar]
- Dubnau E., Weir J., Nair G., Carter L., 3rd, Moran C., Jr, Smith I. Bacillus sporulation gene spo0H codes for sigma 30 (sigma H). J Bacteriol. 1988 Mar;170(3):1054–1062. doi: 10.1128/jb.170.3.1054-1062.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feavers I. M., Price V., Moir A. The regulation of the fumarase (citG) gene of Bacillus subtilis 168. Mol Gen Genet. 1988 Mar;211(3):465–471. doi: 10.1007/BF00425702. [DOI] [PubMed] [Google Scholar]
- Ferrari F. A., Trach K., LeCoq D., Spence J., Ferrari E., Hoch J. A. Characterization of the spo0A locus and its deduced product. Proc Natl Acad Sci U S A. 1985 May;82(9):2647–2651. doi: 10.1073/pnas.82.9.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forst S., Inouye M. Environmentally regulated gene expression for membrane proteins in Escherichia coli. Annu Rev Cell Biol. 1988;4:21–42. doi: 10.1146/annurev.cb.04.110188.000321. [DOI] [PubMed] [Google Scholar]
- Grossman A. D., Losick R. Extracellular control of spore formation in Bacillus subtilis. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4369–4373. doi: 10.1073/pnas.85.12.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healy J., Weir J., Smith I., Losick R. Post-transcriptional control of a sporulation regulatory gene encoding transcription factor sigma H in Bacillus subtilis. Mol Microbiol. 1991 Feb;5(2):477–487. doi: 10.1111/j.1365-2958.1991.tb02131.x. [DOI] [PubMed] [Google Scholar]
- Hess J. F., Bourret R. B., Simon M. I. Histidine phosphorylation and phosphoryl group transfer in bacterial chemotaxis. Nature. 1988 Nov 10;336(6195):139–143. doi: 10.1038/336139a0. [DOI] [PubMed] [Google Scholar]
- Hoch J. A., Mathews J. L. Chromosomal location of pleiotropic negative sporulation mutations in Bacillus subtilis. Genetics. 1973 Feb;73(2):215–228. doi: 10.1093/genetics/73.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt T. P., Magasanik B. Transcription of glnA by purified Escherichia coli components: core RNA polymerase and the products of glnF, glnG, and glnL. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8453–8457. doi: 10.1073/pnas.82.24.8453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igo M. M., Ninfa A. J., Silhavy T. J. A bacterial environmental sensor that functions as a protein kinase and stimulates transcriptional activation. Genes Dev. 1989 May;3(5):598–605. doi: 10.1101/gad.3.5.598. [DOI] [PubMed] [Google Scholar]
- Johnson W. C., Moran C. P., Jr, Losick R. Two RNA polymerase sigma factors from Bacillus subtilis discriminate between overlapping promoters for a developmentally regulated gene. Nature. 1983 Apr 28;302(5911):800–804. doi: 10.1038/302800a0. [DOI] [PubMed] [Google Scholar]
- Kenney T. J., Kirchman P. A., Moran C. P., Jr Gene encoding sigma E is transcribed from a sigma A-like promoter in Bacillus subtilis. J Bacteriol. 1988 Jul;170(7):3058–3064. doi: 10.1128/jb.170.7.3058-3064.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofoid E. C., Parkinson J. S. Transmitter and receiver modules in bacterial signaling proteins. Proc Natl Acad Sci U S A. 1988 Jul;85(14):4981–4985. doi: 10.1073/pnas.85.14.4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudoh J., Ikeuchi T., Kurahashi K. Nucleotide sequences of the sporulation gene spo0A and its mutant genes of Bacillus subtilis. Proc Natl Acad Sci U S A. 1985 May;82(9):2665–2668. doi: 10.1073/pnas.82.9.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandoski M., Dubnau E., Smith I. Transcriptional regulation of the spo0F gene of Bacillus subtilis. J Bacteriol. 1986 Nov;168(2):870–877. doi: 10.1128/jb.168.2.870-877.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick R., Youngman P., Piggot P. J. Genetics of endospore formation in Bacillus subtilis. Annu Rev Genet. 1986;20:625–669. doi: 10.1146/annurev.ge.20.120186.003205. [DOI] [PubMed] [Google Scholar]
- Lukat G. S., McCleary W. R., Stock A. M., Stock J. B. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc Natl Acad Sci U S A. 1992 Jan 15;89(2):718–722. doi: 10.1073/pnas.89.2.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magasanik B. Reversible phosphorylation of an enhancer binding protein regulates the transcription of bacterial nitrogen utilization genes. Trends Biochem Sci. 1988 Dec;13(12):475–479. doi: 10.1016/0968-0004(88)90234-4. [DOI] [PubMed] [Google Scholar]
- Mysliwiec T. H., Errington J., Vaidya A. B., Bramucci M. G. The Bacillus subtilis spo0J gene: evidence for involvement in catabolite repression of sporulation. J Bacteriol. 1991 Mar;173(6):1911–1919. doi: 10.1128/jb.173.6.1911-1919.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninfa A. J., Magasanik B. Covalent modification of the glnG product, NRI, by the glnL product, NRII, regulates the transcription of the glnALG operon in Escherichia coli. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5909–5913. doi: 10.1073/pnas.83.16.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perego M., Cole S. P., Burbulys D., Trach K., Hoch J. A. Characterization of the gene for a protein kinase which phosphorylates the sporulation-regulatory proteins Spo0A and Spo0F of Bacillus subtilis. J Bacteriol. 1989 Nov;171(11):6187–6196. doi: 10.1128/jb.171.11.6187-6196.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perego M., Hoch J. A. Negative regulation of Bacillus subtilis sporulation by the spo0E gene product. J Bacteriol. 1991 Apr;173(8):2514–2520. doi: 10.1128/jb.173.8.2514-2520.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi F. X., Doi R. H. Localization of a second SigH promoter in the Bacillus subtilis sigA operon and regulation of dnaE expression by the promoter. J Bacteriol. 1990 Oct;172(10):5631–5636. doi: 10.1128/jb.172.10.5631-5636.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronson C. W., Nixon B. T., Ausubel F. M. Conserved domains in bacterial regulatory proteins that respond to environmental stimuli. Cell. 1987 Jun 5;49(5):579–581. doi: 10.1016/0092-8674(87)90530-7. [DOI] [PubMed] [Google Scholar]
- Rosenbluh A., Banner C. D., Losick R., Fitz-James P. C. Identification of a new developmental locus in Bacillus subtilis by construction of a deletion mutation in a cloned gene under sporulation control. J Bacteriol. 1981 Oct;148(1):341–351. doi: 10.1128/jb.148.1.341-351.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner D. Z., LeDeaux J. R., Ireton K., Grossman A. D. The spo0K locus of Bacillus subtilis is homologous to the oligopeptide permease locus and is required for sporulation and competence. J Bacteriol. 1991 Feb;173(4):1388–1398. doi: 10.1128/jb.173.4.1388-1398.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders D. A., Gillece-Castro B. L., Stock A. M., Burlingame A. L., Koshland D. E., Jr Identification of the site of phosphorylation of the chemotaxis response regulator protein, CheY. J Biol Chem. 1989 Dec 25;264(36):21770–21778. [PubMed] [Google Scholar]
- Satola S., Kirchman P. A., Moran C. P., Jr Spo0A binds to a promoter used by sigma A RNA polymerase during sporulation in Bacillus subtilis. Proc Natl Acad Sci U S A. 1991 May 15;88(10):4533–4537. doi: 10.1073/pnas.88.10.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer P., Millet J., Aubert J. P. Catabolic repression of bacterial sporulation. Proc Natl Acad Sci U S A. 1965 Sep;54(3):704–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorenstein R. G., Losick R. Comparative size and properties of the sigma subunits of ribonucleic acid polymerase from Bacillus subtilis and Escherichia coli. J Biol Chem. 1973 Sep 10;248(17):6170–6173. [PubMed] [Google Scholar]
- Slauch J. M., Silhavy T. J. Genetic analysis of the switch that controls porin gene expression in Escherichia coli K-12. J Mol Biol. 1989 Nov 20;210(2):281–292. doi: 10.1016/0022-2836(89)90330-6. [DOI] [PubMed] [Google Scholar]
- Stock A. M., Wylie D. C., Mottonen J. M., Lupas A. N., Ninfa E. G., Ninfa A. J., Schutt C. E., Stock J. B. Phosphoproteins involved in bacterial signal transduction. Cold Spring Harb Symp Quant Biol. 1988;53(Pt 1):49–57. doi: 10.1101/sqb.1988.053.01.009. [DOI] [PubMed] [Google Scholar]
- Stock A., Chen T., Welsh D., Stock J. CheA protein, a central regulator of bacterial chemotaxis, belongs to a family of proteins that control gene expression in response to changing environmental conditions. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1403–1407. doi: 10.1073/pnas.85.5.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock J. B., Ninfa A. J., Stock A. M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989 Dec;53(4):450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock J. B., Stock A. M., Mottonen J. M. Signal transduction in bacteria. Nature. 1990 Mar 29;344(6265):395–400. doi: 10.1038/344395a0. [DOI] [PubMed] [Google Scholar]
- Strauch M., Webb V., Spiegelman G., Hoch J. A. The SpoOA protein of Bacillus subtilis is a repressor of the abrB gene. Proc Natl Acad Sci U S A. 1990 Mar;87(5):1801–1805. doi: 10.1073/pnas.87.5.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatti K. M., Carter H. L., 3rd, Moir A., Moran C. P., Jr Sigma H-directed transcription of citG in Bacillus subtilis. J Bacteriol. 1989 Nov;171(11):5928–5932. doi: 10.1128/jb.171.11.5928-5932.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trach K. A., Chapman J. W., Piggot P. J., Hoch J. A. Deduced product of the stage 0 sporulation gene spo0F shares homology with the Spo0A, OmpR, and SfrA proteins. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7260–7264. doi: 10.1073/pnas.82.21.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trach K., Burbulys D., Strauch M., Wu J. J., Dhillon N., Jonas R., Hanstein C., Kallio P., Perego M., Bird T. Control of the initiation of sporulation in Bacillus subtilis by a phosphorelay. Res Microbiol. 1991 Sep-Oct;142(7-8):815–823. doi: 10.1016/0923-2508(91)90060-n. [DOI] [PubMed] [Google Scholar]
- Trach K., Chapman J. W., Piggot P., LeCoq D., Hoch J. A. Complete sequence and transcriptional analysis of the spo0F region of the Bacillus subtilis chromosome. J Bacteriol. 1988 Sep;170(9):4194–4208. doi: 10.1128/jb.170.9.4194-4208.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueshima R., Fujita N., Ishihama A. DNA supercoiling and temperature shift affect the promoter activity of the Escherichia coli rpoH gene encoding the heat-shock sigma subunit of RNA polymerase. Mol Gen Genet. 1989 Jan;215(2):185–189. doi: 10.1007/BF00339716. [DOI] [PubMed] [Google Scholar]
- Weir J., Predich M., Dubnau E., Nair G., Smith I. Regulation of spo0H, a gene coding for the Bacillus subtilis sigma H factor. J Bacteriol. 1991 Jan;173(2):521–529. doi: 10.1128/jb.173.2.521-529.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. J., Howard M. G., Piggot P. J. Regulation of transcription of the Bacillus subtilis spoIIA locus. J Bacteriol. 1989 Feb;171(2):692–698. doi: 10.1128/jb.171.2.692-698.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. J., Piggot P. J., Tatti K. M., Moran C. P., Jr Transcription of the Bacillus subtilis spoIIA locus. Gene. 1991 May 15;101(1):113–116. doi: 10.1016/0378-1119(91)90231-y. [DOI] [PubMed] [Google Scholar]
- Yamashita S., Yoshikawa H., Kawamura F., Takahashi H., Yamamoto T., Kobayashi Y., Saito H. The effect of spo0 mutations on the expression of spo0A- and spo0F-lacZ fusions. Mol Gen Genet. 1986 Oct;205(1):28–33. doi: 10.1007/BF02428029. [DOI] [PubMed] [Google Scholar]
- York K., Kenney T. J., Satola S., Moran C. P., Jr, Poth H., Youngman P. Spo0A controls the sigma A-dependent activation of Bacillus subtilis sporulation-specific transcription unit spoIIE. J Bacteriol. 1992 Apr;174(8):2648–2658. doi: 10.1128/jb.174.8.2648-2658.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa H., Kazami J., Yamashita S., Chibazakura T., Sone H., Kawamura F., Oda M., Isaka M., Kobayashi Y., Saito H. Revised assignment for the Bacillus subtilis spo0F gene and its homology with spo0A and with two Escherichia coli genes. Nucleic Acids Res. 1986 Jan 24;14(2):1063–1072. doi: 10.1093/nar/14.2.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber P., Healy J., Carter H. L., 3rd, Cutting S., Moran C. P., Jr, Losick R. Mutation changing the specificity of an RNA polymerase sigma factor. J Mol Biol. 1989 Apr 20;206(4):605–614. doi: 10.1016/0022-2836(89)90569-x. [DOI] [PubMed] [Google Scholar]
- Zuber P., Losick R. Use of a lacZ fusion to study the role of the spoO genes of Bacillus subtilis in developmental regulation. Cell. 1983 Nov;35(1):275–283. doi: 10.1016/0092-8674(83)90230-1. [DOI] [PubMed] [Google Scholar]








