Abstract
We investigated the role of the umuDC and samAB operons in the UV mutability of Salmonella typhimurium. umuDC is located on the chromosome, whereas samAB resides on the virulence plasmid pSLT. Using allele replacement and plasmid curing techniques, we found that UV mutability was eliminated when any of three different umuDC alleles (umuD1, umuC1, or umuD1 umuC1) were on the chromosome even when samAB was present. We conclude that samAB normally does not complement umuDC function in S. typhimurium.
Full text
PDF

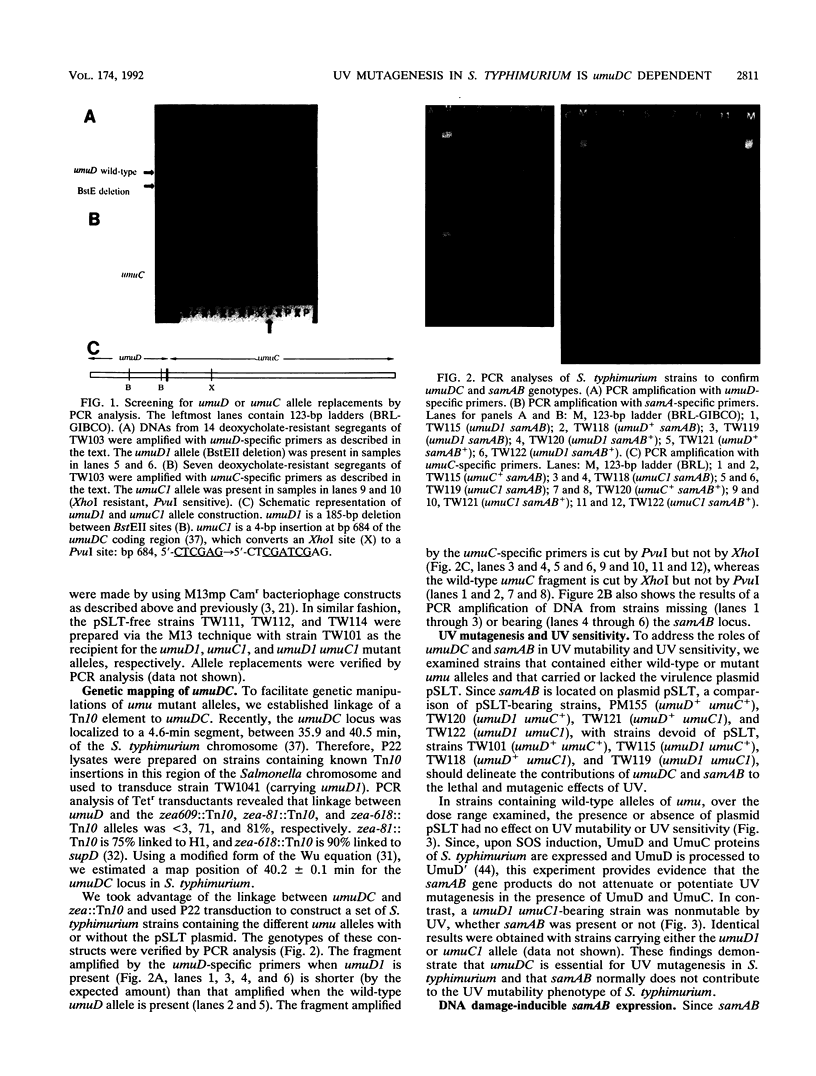
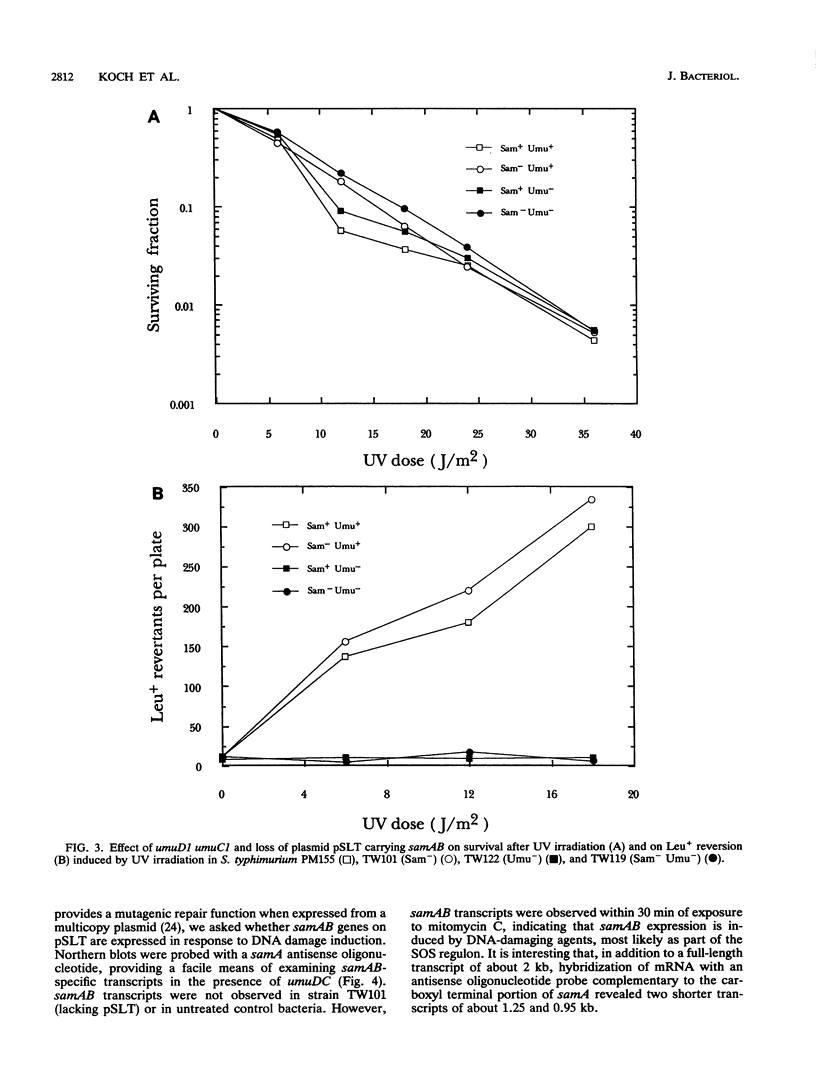



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aiba H., Adhya S., de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem. 1981 Nov 25;256(22):11905–11910. [PubMed] [Google Scholar]
- Blot M., Meyer J., Arber W. Bleomycin-resistance gene derived from the transposon Tn5 confers selective advantage to Escherichia coli K-12. Proc Natl Acad Sci U S A. 1991 Oct 15;88(20):9112–9116. doi: 10.1073/pnas.88.20.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum P., Holzschu D., Kwan H. S., Riggs D., Artz S. Gene replacement and retrieval with recombinant M13mp bacteriophages. J Bacteriol. 1989 Jan;171(1):538–546. doi: 10.1128/jb.171.1.538-546.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner B. R., Huang H. C., Schieven G. L., Ames B. N. Positive selection for loss of tetracycline resistance. J Bacteriol. 1980 Aug;143(2):926–933. doi: 10.1128/jb.143.2.926-933.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calero S., Garriga X., Barbé J. One-step cloning system for isolation of bacterial lexA-like genes. J Bacteriol. 1991 Nov;173(22):7345–7350. doi: 10.1128/jb.173.22.7345-7350.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casse F., Pascal M. C., Chippaux M. Comparison between the chromosomal maps of Escherichia coli and Salmonella typhimurium. Length of the inverted segment in the trp region. Mol Gen Genet. 1973 Aug 17;124(3):253–257. doi: 10.1007/BF00293096. [DOI] [PubMed] [Google Scholar]
- DAVIS B. D., MINGIOLI E. S. Mutants of Escherichia coli requiring methionine or vitamin B12. J Bacteriol. 1950 Jul;60(1):17–28. doi: 10.1128/jb.60.1.17-28.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowden S. B., Strike P. R46-derived recombinant plasmids affecting DNA repair and mutation in E. coli. Mol Gen Genet. 1982;186(1):140–144. doi: 10.1007/BF00422926. [DOI] [PubMed] [Google Scholar]
- Echols H., Goodman M. F. Fidelity mechanisms in DNA replication. Annu Rev Biochem. 1991;60:477–511. doi: 10.1146/annurev.bi.60.070191.002401. [DOI] [PubMed] [Google Scholar]
- Echols H. Mutation rate: some biological and biochemical considerations. Biochimie. 1982 Aug-Sep;64(8-9):571–575. doi: 10.1016/s0300-9084(82)80089-8. [DOI] [PubMed] [Google Scholar]
- Echols H. SOS functions, cancer and inducible evolution. Cell. 1981 Jul;25(1):1–2. doi: 10.1016/0092-8674(81)90223-3. [DOI] [PubMed] [Google Scholar]
- Hill C. W., Gray J. A., Brody H. Use of the isocitrate dehydrogenase structural gene for attachment of e14 in Escherichia coli K-12. J Bacteriol. 1989 Jul;171(7):4083–4084. doi: 10.1128/jb.171.7.4083-4084.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodwick D., Owen D., Strike P. DNA sequence analysis of the imp UV protection and mutation operon of the plasmid TP110: identification of a third gene. Nucleic Acids Res. 1990 Sep 11;18(17):5045–5050. doi: 10.1093/nar/18.17.5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann J., Spingarn N. E., Kobori J., Ames B. N. Detection of carcinogens as mutagens: bacterial tester strains with R factor plasmids. Proc Natl Acad Sci U S A. 1975 Mar;72(3):979–983. doi: 10.1073/pnas.72.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melechen N. E., Go G. Induction of lambdoid prophages by amino acid deprivation: differential inducibility; role of recA. Mol Gen Genet. 1980;180(1):147–155. doi: 10.1007/BF00267364. [DOI] [PubMed] [Google Scholar]
- Miller R. V., Kokjohn T. A. General microbiology of recA: environmental and evolutionary significance. Annu Rev Microbiol. 1990;44:365–394. doi: 10.1146/annurev.mi.44.100190.002053. [DOI] [PubMed] [Google Scholar]
- Mortelmans K. E., Stocker B. A. Ultraviolet light protection, enhancement of ultraviolet light mutagenesis, and mutator effect of plasmid R46 in Salmonella typhimurium. J Bacteriol. 1976 Oct;128(1):271–282. doi: 10.1128/jb.128.1.271-282.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohmi T., Battista J. R., Dodson L. A., Walker G. C. RecA-mediated cleavage activates UmuD for mutagenesis: mechanistic relationship between transcriptional derepression and posttranslational activation. Proc Natl Acad Sci U S A. 1988 Mar;85(6):1816–1820. doi: 10.1073/pnas.85.6.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohmi T., Hakura A., Nakai Y., Watanabe M., Murayama S. Y., Sofuni T. Salmonella typhimurium has two homologous but different umuDC operons: cloning of a new umuDC-like operon (samAB) present in a 60-megadalton cryptic plasmid of S. typhimurium. J Bacteriol. 1991 Feb;173(3):1051–1063. doi: 10.1128/jb.173.3.1051-1063.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orrego C., Eisenstadt E. An inducible pathway is required for mutagenesis in Salmonella typhimurium LT2. J Bacteriol. 1987 Jun;169(6):2885–2888. doi: 10.1128/jb.169.6.2885-2888.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbye K. M., Margolin P. Role of the supX gene in ultraviolet light-induced mutagenesis in Salmonella typhimurium. J Bacteriol. 1981 Apr;146(1):170–178. doi: 10.1128/jb.146.1.170-178.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radman M. Is there SOS induction in mammalian cells? Photochem Photobiol. 1980 Dec;32(6):823–830. doi: 10.1111/j.1751-1097.1980.tb04062.x. [DOI] [PubMed] [Google Scholar]
- Raymond-Denise A., Guillen N. Identification of dinR, a DNA damage-inducible regulator gene of Bacillus subtilis. J Bacteriol. 1991 Nov;173(22):7084–7091. doi: 10.1128/jb.173.22.7084-7091.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson K. E., Roth J. R. Linkage map of Salmonella typhimurium, edition VII. Microbiol Rev. 1988 Dec;52(4):485–532. doi: 10.1128/mr.52.4.485-532.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedgwick S. G., Ho C., Woodgate R. Mutagenic DNA repair in enterobacteria. J Bacteriol. 1991 Sep;173(18):5604–5611. doi: 10.1128/jb.173.18.5604-5611.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba T., Iwasaki H., Nakata A., Shinagawa H. Proteolytic processing of MucA protein in SOS mutagenesis: both processed and unprocessed MucA may be active in the mutagenesis. Mol Gen Genet. 1990 Nov;224(2):169–176. doi: 10.1007/BF00271549. [DOI] [PubMed] [Google Scholar]
- Shinagawa H., Iwasaki H., Kato T., Nakata A. RecA protein-dependent cleavage of UmuD protein and SOS mutagenesis. Proc Natl Acad Sci U S A. 1988 Mar;85(6):1806–1810. doi: 10.1073/pnas.85.6.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slilaty S. N., Little J. W. Lysine-156 and serine-119 are required for LexA repressor cleavage: a possible mechanism. Proc Natl Acad Sci U S A. 1987 Jun;84(12):3987–3991. doi: 10.1073/pnas.84.12.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. M., Koch W. H., Franklin S. B., Foster P. L., Cebula T. A., Eisenstadt E. Sequence analysis and mapping of the Salmonella typhimurium LT2 umuDC operon. J Bacteriol. 1990 Sep;172(9):4964–4978. doi: 10.1128/jb.172.9.4964-4978.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strike P., Lodwick D. Plasmid genes affecting DNA repair and mutation. J Cell Sci Suppl. 1987;6:303–321. doi: 10.1242/jcs.1984.supplement_6.20. [DOI] [PubMed] [Google Scholar]
- Tanooka H., Tanaka K., Shinozaki K. Heterospecific expression of misrepair-enhancing activity of mucAB in Escherichia coli and Bacillus subtilis. J Bacteriol. 1991 May;173(9):2906–2914. doi: 10.1128/jb.173.9.2906-2914.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S. M., Crowne H. M., Pidsley S. C., Sedgwick S. G. Structural characterization of the Salmonella typhimurium LT2 umu operon. J Bacteriol. 1990 Sep;172(9):4979–4987. doi: 10.1128/jb.172.9.4979-4987.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinge S. A., Curtiss R., 3rd Isolation of the replication and partitioning regions of the Salmonella typhimurium virulence plasmid and stabilization of heterologous replicons. J Bacteriol. 1990 Sep;172(9):5266–5277. doi: 10.1128/jb.172.9.5266-5277.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G. C. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Rev. 1984 Mar;48(1):60–93. doi: 10.1128/mr.48.1.60-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodgate R. Construction of a umuDC operon substitution mutation in Escherichia coli. Mutat Res. 1992 Mar;281(3):221–225. doi: 10.1016/0165-7992(92)90012-7. [DOI] [PubMed] [Google Scholar]
- Woodgate R., Levine A. S., Koch W. H., Cebula T. A., Eisenstadt E. Induction and cleavage of Salmonella typhimurium UmuD protein. Mol Gen Genet. 1991 Sep;229(1):81–85. doi: 10.1007/BF00264216. [DOI] [PubMed] [Google Scholar]