Abstract
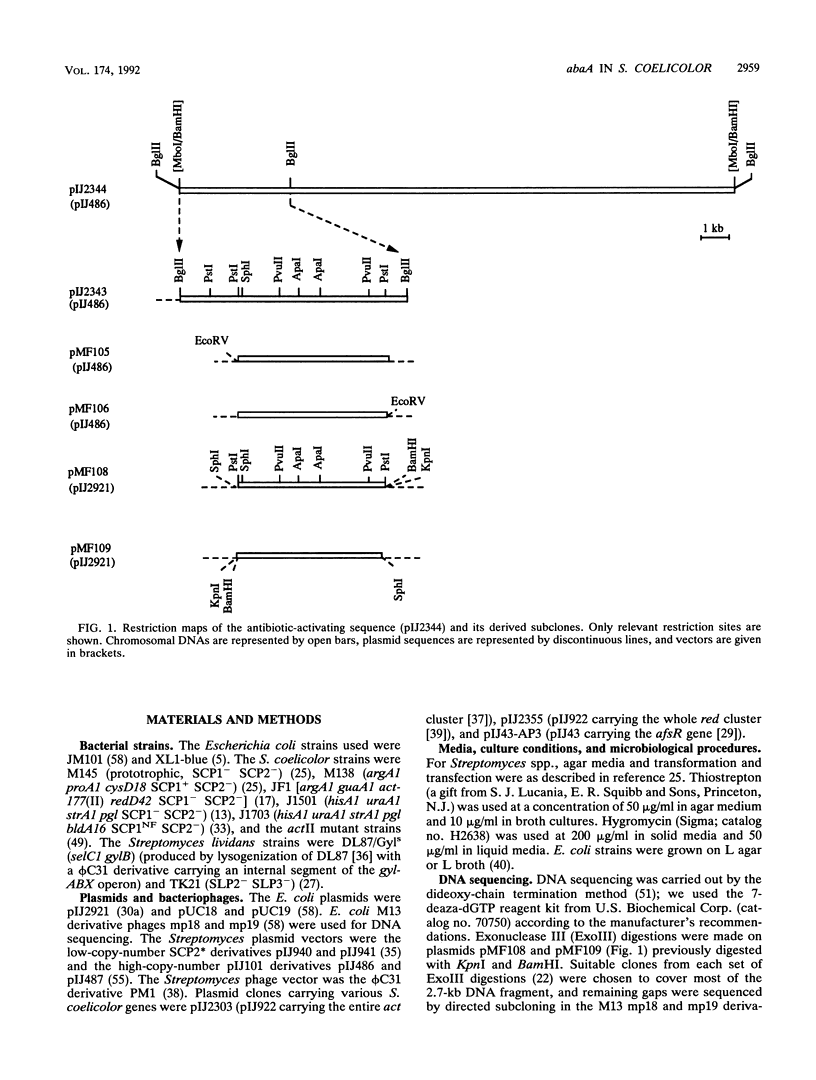
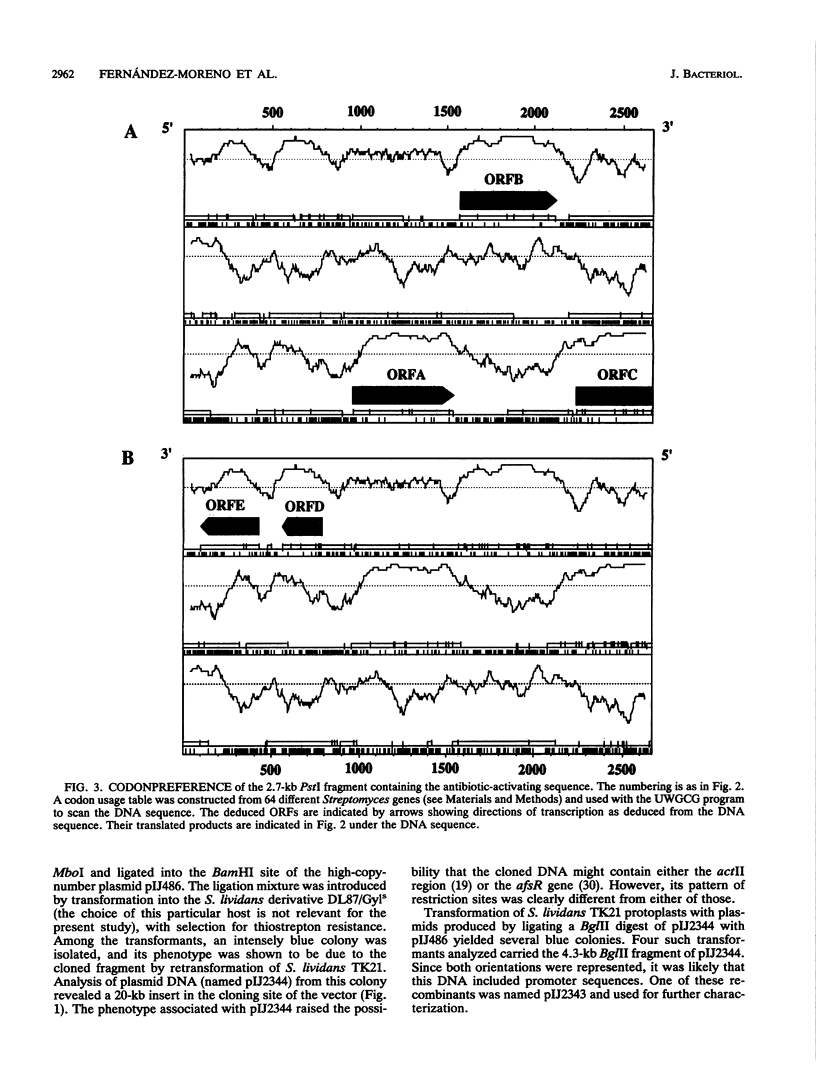
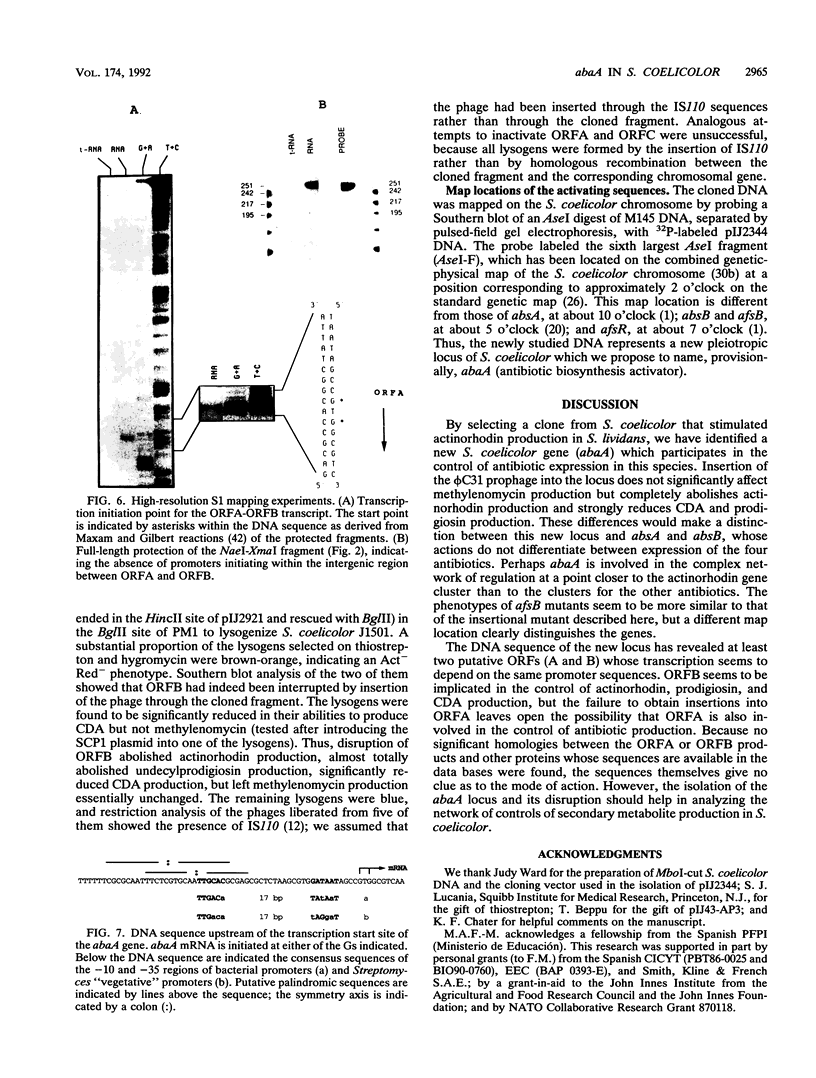
Production of the blue-pigmented antibiotic actinorhodin is greatly enhanced in Streptomyces lividans and Streptomyces coelicolor by transformation with a 2.7-kb DNA fragment from the S. coelicolor chromosome cloned on a multicopy plasmid. Southern analysis, restriction map comparisons, and map locations of the cloned genes revealed that these genes were different from other known S. coelicolor genes concerned with actinorhodin biosynthesis or its pleiotropic regulation. Computer analysis of the DNA sequence showed five putative open reading frames (ORFs), which were named ORFA, ORFB, and ORFC (transcribed in one direction) and ORFD and ORFE (transcribed in the opposite direction). Subcloning experiments revealed that ORFB together with 137 bp downstream of it is responsible for antibiotic overproduction in S. lividans. Insertion of a phi C31 prophage into ORFB by homologous recombination gave rise to a mutant phenotype in which the production of actinorhodin, undecylprodigiosin, and the calcium-dependent antibiotic (but not methylenomycin) was reduced or abolished. The nonproducing mutants were not affected in the timing or vigor or sporulation. A possible involvement of ORFA in antibiotic production in S. coelicolor is not excluded. abaA constitutes a new locus which, like the afs and abs genes previously described, pleiotropically regulates antibiotic production. DNA sequences that hybridize with the cloned DNA are present in several different Streptomyces species.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adamidis T., Riggle P., Champness W. Mutations in a new Streptomyces coelicolor locus which globally block antibiotic biosynthesis but not sporulation. J Bacteriol. 1990 Jun;172(6):2962–2969. doi: 10.1128/jb.172.6.2962-2969.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzai H., Murakami T., Imai S., Satoh A., Nagaoka K., Thompson C. J. Transcriptional regulation of bialaphos biosynthesis in Streptomyces hygroscopicus. J Bacteriol. 1987 Aug;169(8):3482–3488. doi: 10.1128/jb.169.8.3482-3488.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb M. J., Cohen S. N. Gene expression in Streptomyces: construction and application of promoter-probe plasmid vectors in Streptomyces lividans. Mol Gen Genet. 1982;187(2):265–277. doi: 10.1007/BF00331128. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Findlay P. R., Johnson M. W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984 Oct;30(1-3):157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- Chater K. F., Bruton C. J., Foster S. G., Tobek I. Physical and genetic analysis of IS110, a transposable element of Streptomyces coelicolor A3(2). Mol Gen Genet. 1985;200(2):235–239. doi: 10.1007/BF00425429. [DOI] [PubMed] [Google Scholar]
- Chater K. F., Bruton C. J., King A. A., Suarez J. E. The expression of Streptomyces and Escherichia coli drug-resistance determinants cloned into the Streptomyces phage phi C31. Gene. 1982 Jul-Aug;19(1):21–32. doi: 10.1016/0378-1119(82)90185-8. [DOI] [PubMed] [Google Scholar]
- Chater K. F., Bruton C. J. Mutational cloning in Streptomyces and the isolation of antibiotic production genes. Gene. 1983 Dec;26(1):67–78. doi: 10.1016/0378-1119(83)90037-9. [DOI] [PubMed] [Google Scholar]
- Chater K. F., Bruton C. J. Resistance, regulatory and production genes for the antibiotic methylenomycin are clustered. EMBO J. 1985 Jul;4(7):1893–1897. doi: 10.1002/j.1460-2075.1985.tb03866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chater K. F. Multilevel regulation of Streptomyces differentiation. Trends Genet. 1989 Nov;5(11):372–377. doi: 10.1016/0168-9525(89)90172-8. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distler J., Ebert A., Mansouri K., Pissowotzki K., Stockmann M., Piepersberg W. Gene cluster for streptomycin biosynthesis in Streptomyces griseus: nucleotide sequence of three genes and analysis of transcriptional activity. Nucleic Acids Res. 1987 Oct 12;15(19):8041–8056. doi: 10.1093/nar/15.19.8041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd I. B., Egan J. B. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 1990 Sep 11;18(17):5019–5026. doi: 10.1093/nar/18.17.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feitelson J. S., Hopwood D. A. Cloning of a Streptomyces gene for an O-methyltransferase involved in antibiotic biosynthesis. Mol Gen Genet. 1983;190(3):394–398. doi: 10.1007/BF00331065. [DOI] [PubMed] [Google Scholar]
- Feitelson J. S., Malpartida F., Hopwood D. A. Genetic and biochemical characterization of the red gene cluster of Streptomyces coelicolor A3(2). J Gen Microbiol. 1985 Sep;131(9):2431–2441. doi: 10.1099/00221287-131-9-2431. [DOI] [PubMed] [Google Scholar]
- Fernández-Moreno M. A., Caballero J. L., Hopwood D. A., Malpartida F. The act cluster contains regulatory and antibiotic export genes, direct targets for translational control by the bldA tRNA gene of Streptomyces. Cell. 1991 Aug 23;66(4):769–780. doi: 10.1016/0092-8674(91)90120-n. [DOI] [PubMed] [Google Scholar]
- Hara O., Horinouchi S., Uozumi T., Beppu T. Genetic analysis of A-factor synthesis in Streptomyces coelicolor A3(2) and Streptomyces griseus. J Gen Microbiol. 1983 Sep;129(9):2939–2944. doi: 10.1099/00221287-129-9-2939. [DOI] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Hopwood D. A., Kieser T., Wright H. M., Bibb M. J. Plasmids, recombination and chromosome mapping in Streptomyces lividans 66. J Gen Microbiol. 1983 Jul;129(7):2257–2269. doi: 10.1099/00221287-129-7-2257. [DOI] [PubMed] [Google Scholar]
- Hopwood D. A. The Leeuwenhoek lecture, 1987. Towards an understanding of gene switching in Streptomyces, the basis of sporulation and antibiotic production. Proc R Soc Lond B Biol Sci. 1988 Nov 22;235(1279):121–138. doi: 10.1098/rspb.1988.0067. [DOI] [PubMed] [Google Scholar]
- Hopwood D. A., Wright H. M. CDA is a new chromosomally-determined antibiotic from Streptomyces coelicolor A3(2). J Gen Microbiol. 1983 Dec;129(12):3575–3579. doi: 10.1099/00221287-129-12-3575. [DOI] [PubMed] [Google Scholar]
- Horinouchi S., Hara O., Beppu T. Cloning of a pleiotropic gene that positively controls biosynthesis of A-factor, actinorhodin, and prodigiosin in Streptomyces coelicolor A3(2) and Streptomyces lividans. J Bacteriol. 1983 Sep;155(3):1238–1248. doi: 10.1128/jb.155.3.1238-1248.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Kito M., Nishiyama M., Furuya K., Hong S. K., Miyake K., Beppu T. Primary structure of AfsR, a global regulatory protein for secondary metabolite formation in Streptomyces coelicolor A3(2). Gene. 1990 Oct 30;95(1):49–56. doi: 10.1016/0378-1119(90)90412-k. [DOI] [PubMed] [Google Scholar]
- Lakey J. H., Lea E. J., Rudd B. A., Wright H. M., Hopwood D. A. A new channel-forming antibiotic from Streptomyces coelicolor A3(2) which requires calcium for its activity. J Gen Microbiol. 1983 Dec;129(12):3565–3573. doi: 10.1099/00221287-129-12-3565. [DOI] [PubMed] [Google Scholar]
- Lanzer M., Bujard H. Promoters largely determine the efficiency of repressor action. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8973–8977. doi: 10.1073/pnas.85.23.8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor E. J., Baylis H. A., Chater K. F. Pleiotropic morphological and antibiotic deficiencies result from mutations in a gene encoding a tRNA-like product in Streptomyces coelicolor A3(2). Genes Dev. 1987 Dec;1(10):1305–1310. doi: 10.1101/gad.1.10.1305. [DOI] [PubMed] [Google Scholar]
- Leskiw B. K., Lawlor E. J., Fernandez-Abalos J. M., Chater K. F. TTA codons in some genes prevent their expression in a class of developmental, antibiotic-negative, Streptomyces mutants. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2461–2465. doi: 10.1073/pnas.88.6.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydiate D. J., Malpartida F., Hopwood D. A. The Streptomyces plasmid SCP2*: its functional analysis and development into useful cloning vectors. Gene. 1985;35(3):223–235. doi: 10.1016/0378-1119(85)90001-0. [DOI] [PubMed] [Google Scholar]
- Malpartida F., Hopwood D. A. Molecular cloning of the whole biosynthetic pathway of a Streptomyces antibiotic and its expression in a heterologous host. 1984 May 31-Jun 6Nature. 309(5967):462–464. doi: 10.1038/309462a0. [DOI] [PubMed] [Google Scholar]
- Malpartida F., Hopwood D. A. Physical and genetic characterisation of the gene cluster for the antibiotic actinorhodin in Streptomyces coelicolor A3(2). Mol Gen Genet. 1986 Oct;205(1):66–73. doi: 10.1007/BF02428033. [DOI] [PubMed] [Google Scholar]
- Malpartida F., Niemi J., Navarrete R., Hopwood D. A. Cloning and expression in a heterologous host of the complete set of genes for biosynthesis of the Streptomyces coelicolor antibiotic undecylprodigiosin. Gene. 1990 Sep 1;93(1):91–99. doi: 10.1016/0378-1119(90)90141-d. [DOI] [PubMed] [Google Scholar]
- Martín M. F., Liras P. Organization and expression of genes involved in the biosynthesis of antibiotics and other secondary metabolites. Annu Rev Microbiol. 1989;43:173–206. doi: 10.1146/annurev.mi.43.100189.001133. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Murray M. G. Use of sodium trichloroacetate and mung bean nuclease to increase sensitivity and precision during transcript mapping. Anal Biochem. 1986 Oct;158(1):165–170. doi: 10.1016/0003-2697(86)90605-6. [DOI] [PubMed] [Google Scholar]
- Narva K. E., Feitelson J. S. Nucleotide sequence and transcriptional analysis of the redD locus of Streptomyces coelicolor A3(2). J Bacteriol. 1990 Jan;172(1):326–333. doi: 10.1128/jb.172.1.326-333.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnuki T., Imanaka T., Aiba S. Self-cloning in Streptomyces griseus of an str gene cluster for streptomycin biosynthesis and streptomycin resistance. J Bacteriol. 1985 Oct;164(1):85–94. doi: 10.1128/jb.164.1.85-94.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oki T., Matsuzawa Y., Kiyoshima K., Yoshimoto A., Naganawa H., Takeuchi T., Umezawa H. New anthracyclines, feudomycins, produced by the mutant from Streptomyces coeruleorubidus ME130-A4. J Antibiot (Tokyo) 1981 Jul;34(7):783–790. doi: 10.7164/antibiotics.34.783. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Rudd B. A., Hopwood D. A. A pigmented mycelial antibiotic in Streptomyces coelicolor: control by a chromosomal gene cluster. J Gen Microbiol. 1980 Aug;119(2):333–340. doi: 10.1099/00221287-119-2-333. [DOI] [PubMed] [Google Scholar]
- Rudd B. A., Hopwood D. A. Genetics of actinorhodin biosynthesis by Streptomyces coelicolor A3(2). J Gen Microbiol. 1979 Sep;114(1):35–43. doi: 10.1099/00221287-114-1-35. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer R. T., Yocum R. R., Doolittle R. F., Lewis M., Pabo C. O. Homology among DNA-binding proteins suggests use of a conserved super-secondary structure. Nature. 1982 Jul 29;298(5873):447–451. doi: 10.1038/298447a0. [DOI] [PubMed] [Google Scholar]
- Stein D., Cohen S. N. A cloned regulatory gene of Streptomyces lividans can suppress the pigment deficiency phenotype of different developmental mutants. J Bacteriol. 1989 Apr;171(4):2258–2261. doi: 10.1128/jb.171.4.2258-2261.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. M., Janssen G. R., Kieser T., Bibb M. J., Buttner M. J., Bibb M. J. Construction and characterisation of a series of multi-copy promoter-probe plasmid vectors for Streptomyces using the aminoglycoside phosphotransferase gene from Tn5 as indicator. Mol Gen Genet. 1986 Jun;203(3):468–478. doi: 10.1007/BF00422072. [DOI] [PubMed] [Google Scholar]
- Wright L. F., Hopwood D. A. Actinorhodin is a chromosomally-determined antibiotic in Streptomyces coelicolar A3(2). J Gen Microbiol. 1976 Oct;96(2):289–297. doi: 10.1099/00221287-96-2-289. [DOI] [PubMed] [Google Scholar]
- Wright L. F., Hopwood D. A. Identification of the antibiotic determined by the SCP1 plasmid of Streptomyces coelicolor A3(2). J Gen Microbiol. 1976 Jul;95(1):96–106. doi: 10.1099/00221287-95-1-96. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]




