Abstract
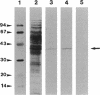
Assessment of potential risks involved in the release of genetically engineered microorganisms is facilitated by the availability of monoclonal antibodies (MAbs), a tool potentially able to monitor specific organisms. We raised a bank of MAbs against the soil bacterium Pseudomonas putida 2440, which is a host for modified TOL plasmids and other recombinant plasmids. Three MAbs, 7.3B, 7.4D, and 7.5D, were highly specific and recognized only P. putida bacteria. Furthermore, we developed a semiquantitative dot blot assay that allowed us to detect as few as 100 cells per spot. A 40-kDa cell surface protein was the target for MAbs 7.4D and 7.5D. Detection of the cell antigen depended on the bacterial growth phase and culture medium. The O antigen of lipopolysaccharide seems to be the target for MAb 7.3B, and its in vivo detection was independent of the bacterial growth phase and culture medium. MAb 7.3B was used successfully to track P. putida (pWW0) released in unsterile lake mesocosms.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abril M. A., Michan C., Timmis K. N., Ramos J. L. Regulator and enzyme specificities of the TOL plasmid-encoded upper pathway for degradation of aromatic hydrocarbons and expansion of the substrate range of the pathway. J Bacteriol. 1989 Dec;171(12):6782–6790. doi: 10.1128/jb.171.12.6782-6790.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R. I., Binder B. J., Olson R. J., Chisholm S. W., Devereux R., Stahl D. A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990 Jun;56(6):1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R. I., Krumholz L., Stahl D. A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990 Feb;172(2):762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagdasarian M., Timmis K. N. Host: vector systems for gene cloning in Pseudomonas. Curr Top Microbiol Immunol. 1982;96:47–67. doi: 10.1007/978-3-642-68315-2_4. [DOI] [PubMed] [Google Scholar]
- Beringer J. E. R factor transfer in Rhizobium leguminosarum. J Gen Microbiol. 1974 Sep;84(1):188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- Carlson R. W. Heterogeneity of Rhizobium lipopolysaccharides. J Bacteriol. 1984 Jun;158(3):1012–1017. doi: 10.1128/jb.158.3.1012-1017.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadesús J., Olivares J. Rough and fine linkage mapping of the Rhizobium meliloti chromosome. Mol Gen Genet. 1979 Jul 13;174(2):203–209. doi: 10.1007/BF00268356. [DOI] [PubMed] [Google Scholar]
- Delgado A., Wubbolts M. G., Abril M. A., Ramos J. L. Nitroaromatics Are Substrates for the TOL Plasmid Upper-Pathway Enzymes. Appl Environ Microbiol. 1992 Jan;58(1):415–417. doi: 10.1128/aem.58.1.415-417.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn E., Hellwig M., Reineke W., Knackmuss H. J. Isolation and characterization of a 3-chlorobenzoate degrading pseudomonad. Arch Microbiol. 1974;99(1):61–70. doi: 10.1007/BF00696222. [DOI] [PubMed] [Google Scholar]
- Fink J. M., Zissler J. F. Characterization of lipopolysaccharide from Myxococcus xanthus by use of monoclonal antibodies. J Bacteriol. 1989 Apr;171(4):2028–2032. doi: 10.1128/jb.171.4.2028-2032.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin F. C., Bagdasarian M., Bagdasarian M. M., Timmis K. N. Molecular and functional analysis of the TOL plasmid pWWO from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta cleavage pathway. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7458–7462. doi: 10.1073/pnas.78.12.7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman R. C., Leive L. Heterogeneity of antigenic-side-chain length in lipopolysaccharide from Escherichia coli 0111 and Salmonella typhimurium LT2. Eur J Biochem. 1980;107(1):145–153. doi: 10.1111/j.1432-1033.1980.tb04635.x. [DOI] [PubMed] [Google Scholar]
- Guyon P., Chilton M. D., Petit A., Tempé J. Agropine in "null-type" crown gall tumors: Evidence for generality of the opine concept. Proc Natl Acad Sci U S A. 1980 May;77(5):2693–2697. doi: 10.1073/pnas.77.5.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchcock P. J., Brown T. M. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol. 1983 Apr;154(1):269–277. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H. C., Schuurink R., Denny T. P., Atkinson M. M., Baker C. J., Yucel I., Hutcheson S. W., Collmer A. Molecular cloning of a Pseudomonas syringae pv. syringae gene cluster that enables Pseudomonas fluorescens to elicit the hypersensitive response in tobacco plants. J Bacteriol. 1988 Oct;170(10):4748–4756. doi: 10.1128/jb.170.10.4748-4756.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jann B., Reske K., Jann K. Heterogeneity of lipopolysaccharides. Analysis of polysaccharide chain lengths by sodium dodecylsulfate-polyacrylamide gel electrophoresis. Eur J Biochem. 1975 Dec 1;60(1):239–246. doi: 10.1111/j.1432-1033.1975.tb20996.x. [DOI] [PubMed] [Google Scholar]
- Kooistra J., Vosman B., Venema G. Cloning and characterization of a Bacillus subtilis transcription unit involved in ATP-dependent DNase synthesis. J Bacteriol. 1988 Oct;170(10):4791–4797. doi: 10.1128/jb.170.10.4791-4797.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler G., Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975 Aug 7;256(5517):495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lanier L. L., Warner N. L. Paraformaldehyde fixation of hematopoietic cells for quantitative flow cytometry (FACS) analysis. J Immunol Methods. 1981;47(1):25–30. doi: 10.1016/0022-1759(81)90253-2. [DOI] [PubMed] [Google Scholar]
- Morgan J. A., Winstanley C., Pickup R. W., Saunders J. R. Rapid Immunocapture of Pseudomonas putida Cells from Lake Water by Using Bacterial Flagella. Appl Environ Microbiol. 1991 Feb;57(2):503–509. doi: 10.1128/aem.57.2.503-509.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandapalan N., Chang B. J. Production and characterization of monoclonal antibodies to Aeromonas sobria surface antigens. FEMS Microbiol Immunol. 1989 Dec;1(8-9):515–524. doi: 10.1111/j.1574-6968.1989.tb02444.x. [DOI] [PubMed] [Google Scholar]
- Neidle E. L., Hartnett C., Bonitz S., Ornston L. N. DNA sequence of the Acinetobacter calcoaceticus catechol 1,2-dioxygenase I structural gene catA: evidence for evolutionary divergence of intradiol dioxygenases by acquisition of DNA sequence repetitions. J Bacteriol. 1988 Oct;170(10):4874–4880. doi: 10.1128/jb.170.10.4874-4880.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop A. J., Bender R. A. RNA polymerase as a repressor of transcription in the hut(P) region of mutant Klebsiella aerogenes histidine utilization operons. J Bacteriol. 1988 Oct;170(10):4986–4990. doi: 10.1128/jb.170.10.4986-4990.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raibaud O., Schwartz M. Positive control of transcription initiation in bacteria. Annu Rev Genet. 1984;18:173–206. doi: 10.1146/annurev.ge.18.120184.001133. [DOI] [PubMed] [Google Scholar]
- Ramos J. L., Timmis K. N. Experimental evolution of catabolic pathways of bacteria. Microbiol Sci. 1987 Aug;4(8):228–237. [PubMed] [Google Scholar]
- Ramos J. L., Wasserfallen A., Rose K., Timmis K. N. Redesigning metabolic routes: manipulation of TOL plasmid pathway for catabolism of alkylbenzoates. Science. 1987 Jan 30;235(4788):593–596. doi: 10.1126/science.3468623. [DOI] [PubMed] [Google Scholar]
- Rivera M., McGroarty E. J. Analysis of a common-antigen lipopolysaccharide from Pseudomonas aeruginosa. J Bacteriol. 1989 Apr;171(4):2244–2248. doi: 10.1128/jb.171.4.2244-2248.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan J. M., Conrad H. E. Structural heterogeneity in the lipopolysaccharide of Salmonella newington. Arch Biochem Biophys. 1974 Jun;162(2):530–535. doi: 10.1016/0003-9861(74)90213-6. [DOI] [PubMed] [Google Scholar]
- Sen K., Nikaido H. Lipopolysaccharide structure required for in vitro trimerization of Escherichia coli OmpF porin. J Bacteriol. 1991 Jan;173(2):926–928. doi: 10.1128/jb.173.2.926-928.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S. J., Gold S., Robeson M., Manulis S., Keen N. T. Structure and organization of the pel genes from Erwinia chrysanthemi EC16. J Bacteriol. 1988 Aug;170(8):3468–3478. doi: 10.1128/jb.170.8.3468-3478.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viebrock A., Zumft W. G. Molecular cloning, heterologous expression, and primary structure of the structural gene for the copper enzyme nitrous oxide reductase from denitrifying Pseudomonas stutzeri. J Bacteriol. 1988 Oct;170(10):4658–4668. doi: 10.1128/jb.170.10.4658-4668.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C. M. Studies and critique of Amido Black 10B, Coomassie Blue R, and Fast Green FCF as stains for proteins after polyacrylamide gel electrophoresis. Anal Biochem. 1979 Jul 15;96(2):263–278. doi: 10.1016/0003-2697(79)90581-5. [DOI] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsey M. J., Williams P. A. Metabolism of toluene and xylenes by Pseudomonas (putida (arvilla) mt-2: evidence for a new function of the TOL plasmid. J Bacteriol. 1975 Oct;124(1):7–13. doi: 10.1128/jb.124.1.7-13.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]