Abstract
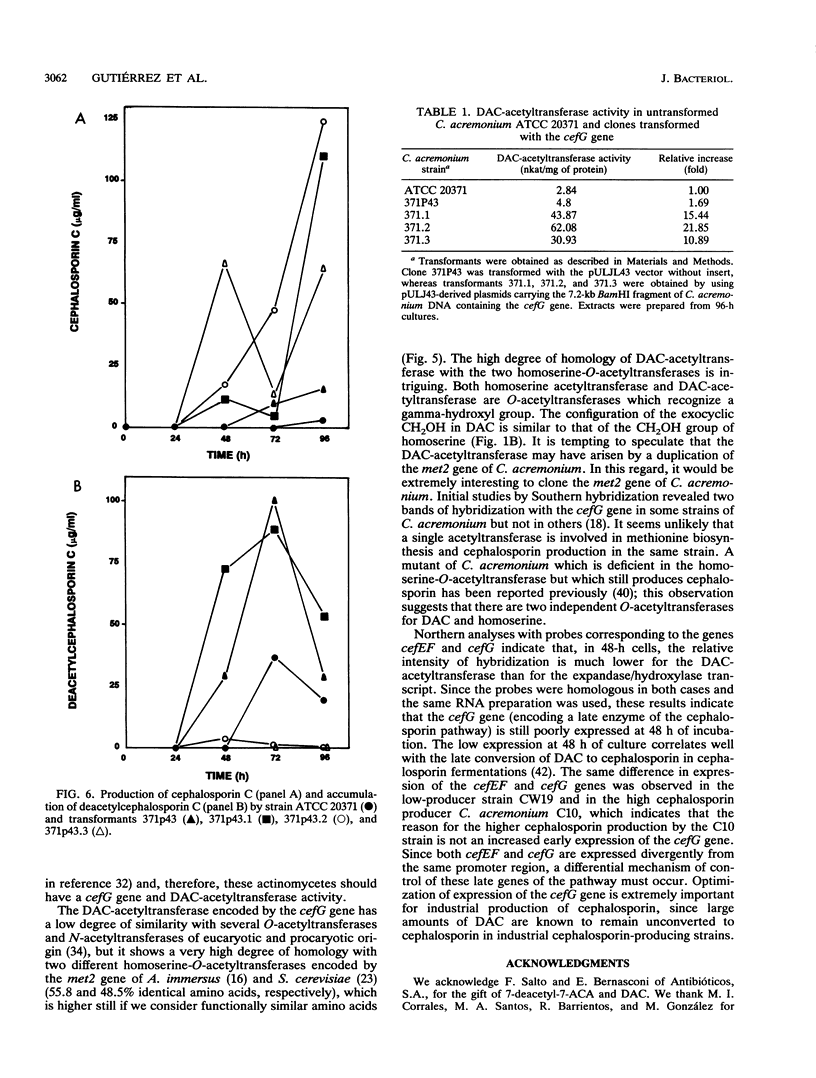
The gene (cefG) encoding the acetyl coenzyme A:deacetylcephalosporin C acetyltransferase of Cephalosporium acremonium (synonym Acremonium chrysogenum) C10 has been cloned. It contains two introns and encodes a protein of 444 amino acids with an M(r) of 49,269 that correlates well with the M(r) deduced by gel filtration. The cefG gene is linked to the cefEF gene (encoding the bifunctional deacetoxycephalosporin C synthase/hydroxylase), but it is expressed in an orientation opposite that of the cefEF gene. Two transcripts of 1.2 and 1.4 kb were found in C. acremonium that correspond to the cefEF and cefG genes, respectively; the degree of expression of the cefG gene was clearly lower than that of the cefEF gene in 48-h cultures. The cloned cefG complemented the deficiency of deacetylcephalosporin acetyltransferase in the nonproducer mutant C. acremonium ATCC 20371 and restored cephalosporin biosynthesis in this strain. Heterologous expression of the cefG genes took place in Penicillium chrysogenum. The deacetylcephalosporin acetyltransferase showed a much higher degree of homology with the O-acetylhomoserine acetyltransferases of Saccharomyces cerevisiae and Ascobolus immersus than with other O-acetyltransferases. The cefEF-cefG cluster of genes encodes the enzymes that carry out the three late steps of the cephalosporin biosynthetic pathway and is not linked to the pcbAB-pcbC gene cluster that encodes the first two steps of the pathway.
Full text
PDF
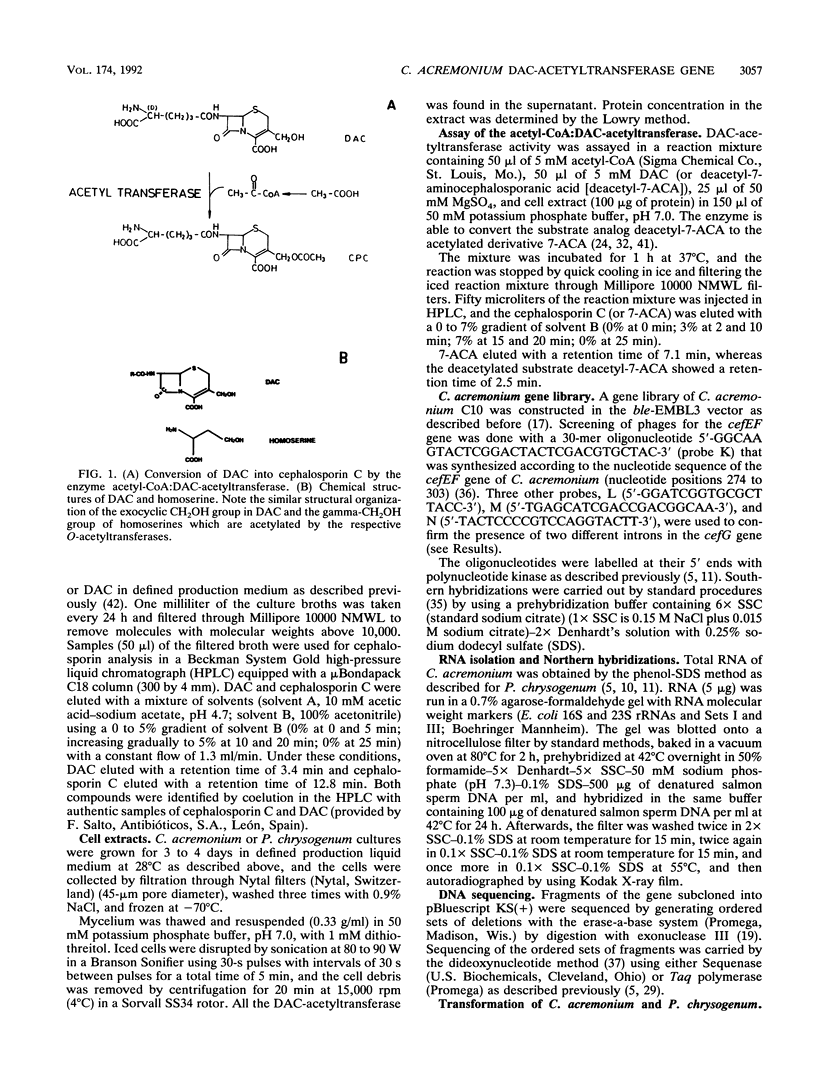




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baldwin J. E., Bird J. W., Field R. A., O'Callaghan N. M., Schofield C. J. Isolation and partial characterisation of ACV synthetase from Cephalosporium acremonium and Streptomyces clavuligerus. J Antibiot (Tokyo) 1990 Aug;43(8):1055–1057. doi: 10.7164/antibiotics.43.1055. [DOI] [PubMed] [Google Scholar]
- Ballance D. J. Sequences important for gene expression in filamentous fungi. Yeast. 1986 Dec;2(4):229–236. doi: 10.1002/yea.320020404. [DOI] [PubMed] [Google Scholar]
- Banko G., Wolfe S., Demain A. L. Cell-free synthesis of delta-(L-alpha-aminoadipyl)-L-cysteine, the first intermediate of penicillin and cephalosporin biosynthesis. Biochem Biophys Res Commun. 1986 May 29;137(1):528–535. doi: 10.1016/0006-291x(86)91242-8. [DOI] [PubMed] [Google Scholar]
- Barredo J. L., Cantoral J. M., Alvarez E., Díez B., Martín J. F. Cloning, sequence analysis and transcriptional study of the isopenicillin N synthase of Penicillium chrysogenum AS-P-78. Mol Gen Genet. 1989 Mar;216(1):91–98. doi: 10.1007/BF00332235. [DOI] [PubMed] [Google Scholar]
- Brannon D. R., Fukuda D. S., Mabe J. A., Huber F. M., Whitney J. G. Detection of a cephalosporin C acetyl esterase in the carbamate cephalosporin antibiotic-producing culture, Streptomyces clavuligerus. Antimicrob Agents Chemother. 1972 Mar;1(3):237–241. doi: 10.1128/aac.1.3.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coque J. J., Liras P., Laiz L., Martín J. F. A gene encoding lysine 6-aminotransferase, which forms the beta-lactam precursor alpha-aminoadipic acid, is located in the cluster of cephamycin biosynthetic genes in Nocardia lactamdurans. J Bacteriol. 1991 Oct;173(19):6258–6264. doi: 10.1128/jb.173.19.6258-6264.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coque J. J., Martín J. F., Calzada J. G., Liras P. The cephamycin biosynthetic genes pcbAB, encoding a large multidomain peptide synthetase, and pcbC of Nocardia lactamdurans are clustered together in an organization different from the same genes in Acremonium chrysogenum and Penicillium chrysogenum. Mol Microbiol. 1991 May;5(5):1125–1133. doi: 10.1111/j.1365-2958.1991.tb01885.x. [DOI] [PubMed] [Google Scholar]
- Dotzlaf J. E., Yeh W. K. Copurification and characterization of deacetoxycephalosporin C synthetase/hydroxylase from Cephalosporium acremonium. J Bacteriol. 1987 Apr;169(4):1611–1618. doi: 10.1128/jb.169.4.1611-1618.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díez B., Barredo J. L., Alvarez E., Cantoral J. M., van Solingen P., Groenen M. A., Veenstra A. E., Martín J. F. Two genes involved in penicillin biosynthesis are linked in a 5.1 kb SalI fragment in the genome of Penicillium chrysogenum. Mol Gen Genet. 1989 Sep;218(3):572–576. doi: 10.1007/BF00332426. [DOI] [PubMed] [Google Scholar]
- Díez B., Gutiérrez S., Barredo J. L., van Solingen P., van der Voort L. H., Martín J. F. The cluster of penicillin biosynthetic genes. Identification and characterization of the pcbAB gene encoding the alpha-aminoadipyl-cysteinyl-valine synthetase and linkage to the pcbC and penDE genes. J Biol Chem. 1990 Sep 25;265(27):16358–16365. [PubMed] [Google Scholar]
- Goyon C., Faugeron G., Rossignol J. L. Molecular cloning and characterization of the met2 gene from Ascobolus immersus. Gene. 1988 Mar 31;63(2):297–308. doi: 10.1016/0378-1119(88)90533-1. [DOI] [PubMed] [Google Scholar]
- Gutiérrez S., Díez B., Montenegro E., Martín J. F. Characterization of the Cephalosporium acremonium pcbAB gene encoding alpha-aminoadipyl-cysteinyl-valine synthetase, a large multidomain peptide synthetase: linkage to the pcbC gene as a cluster of early cephalosporin biosynthetic genes and evidence of multiple functional domains. J Bacteriol. 1991 Apr;173(7):2354–2365. doi: 10.1128/jb.173.7.2354-2365.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Higgens C. E., Hamill R. L., Sands T. H., Hoehn M. M., Davis N. E. Letter: The occurrence of deacetoxycephalosporin C in fungi and streptomycetes. J Antibiot (Tokyo) 1974 Apr;27(4):298–300. doi: 10.7164/antibiotics.27.298. [DOI] [PubMed] [Google Scholar]
- Hoskins J. A., O'Callaghan N., Queener S. W., Cantwell C. A., Wood J. S., Chen V. J., Skatrud P. L. Gene disruption of the pcbAB gene encoding ACV synthetase in Cephalosporium acremonium. Curr Genet. 1990 Dec;18(6):523–530. doi: 10.1007/BF00327023. [DOI] [PubMed] [Google Scholar]
- Kovacevic S., Miller J. R. Cloning and sequencing of the beta-lactam hydroxylase gene (cefF) from Streptomyces clavuligerus: gene duplication may have led to separate hydroxylase and expandase activities in the actinomycetes. J Bacteriol. 1991 Jan;173(1):398–400. doi: 10.1128/jb.173.1.398-400.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langin T., Faugeron G., Goyon C., Nicolas A., Rossignol J. L. The MET2 gene of Saccharomyces cerevisiae: molecular cloning and nucleotide sequence. Gene. 1986;49(3):283–293. doi: 10.1016/0378-1119(86)90364-1. [DOI] [PubMed] [Google Scholar]
- Martin-Zanca D. M., Martín J. F. Carbon catabolite regulation of the conversion of penicillin N into cephalosporin C. J Antibiot (Tokyo) 1983 Jun;36(6):700–708. doi: 10.7164/antibiotics.36.700. [DOI] [PubMed] [Google Scholar]
- Martín M. F., Liras P. Organization and expression of genes involved in the biosynthesis of antibiotics and other secondary metabolites. Annu Rev Microbiol. 1989;43:173–206. doi: 10.1146/annurev.mi.43.100189.001133. [DOI] [PubMed] [Google Scholar]
- Montenegro E., Barredo J. L., Gutiérrez S., Díez B., Alvarez E., Martín J. F. Cloning, characterization of the acyl-CoA:6-amino penicillanic acid acyltransferase gene of Aspergillus nidulans and linkage to the isopenicillin N synthase gene. Mol Gen Genet. 1990 May;221(3):322–330. doi: 10.1007/BF00259395. [DOI] [PubMed] [Google Scholar]
- Nagarajan R., Boeck L. D., Gorman M., Hamill R. L., Higgens C. E., Hoehn M. M., Stark W. M., Whitney J. G. Beta-lactam antibiotics from Streptomyces. J Am Chem Soc. 1971 May 5;93(9):2308–2310. doi: 10.1021/ja00738a035. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J. Eukaryotic promoters? Nature. 1979 May 31;279(5712):376–376. doi: 10.1038/279376a0. [DOI] [PubMed] [Google Scholar]
- Salauze D., Perez-Gonzalez J. A., Piepersberg W., Davies J. Characterisation of aminoglycoside acetyltransferase-encoding genes of neomycin-producing Micromonospora chalcea and Streptomyces fradiae. Gene. 1991 May 15;101(1):143–148. doi: 10.1016/0378-1119(91)90237-6. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheidegger A., Küenzi M. T., Nüesch J. Partial purification and catalytic properties of a bifunctional enzyme in the biosynthetic pathway of beta-lactams in Cephalosporium acremonium. J Antibiot (Tokyo) 1984 May;37(5):522–531. doi: 10.7164/antibiotics.37.522. [DOI] [PubMed] [Google Scholar]
- Skatrud P. L., Queener S. W. An electrophoretic molecular karyotype for an industrial strain of Cephalosporium acremonium. Gene. 1989 May 30;78(2):331–338. doi: 10.1016/0378-1119(89)90235-7. [DOI] [PubMed] [Google Scholar]