Abstract
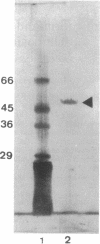
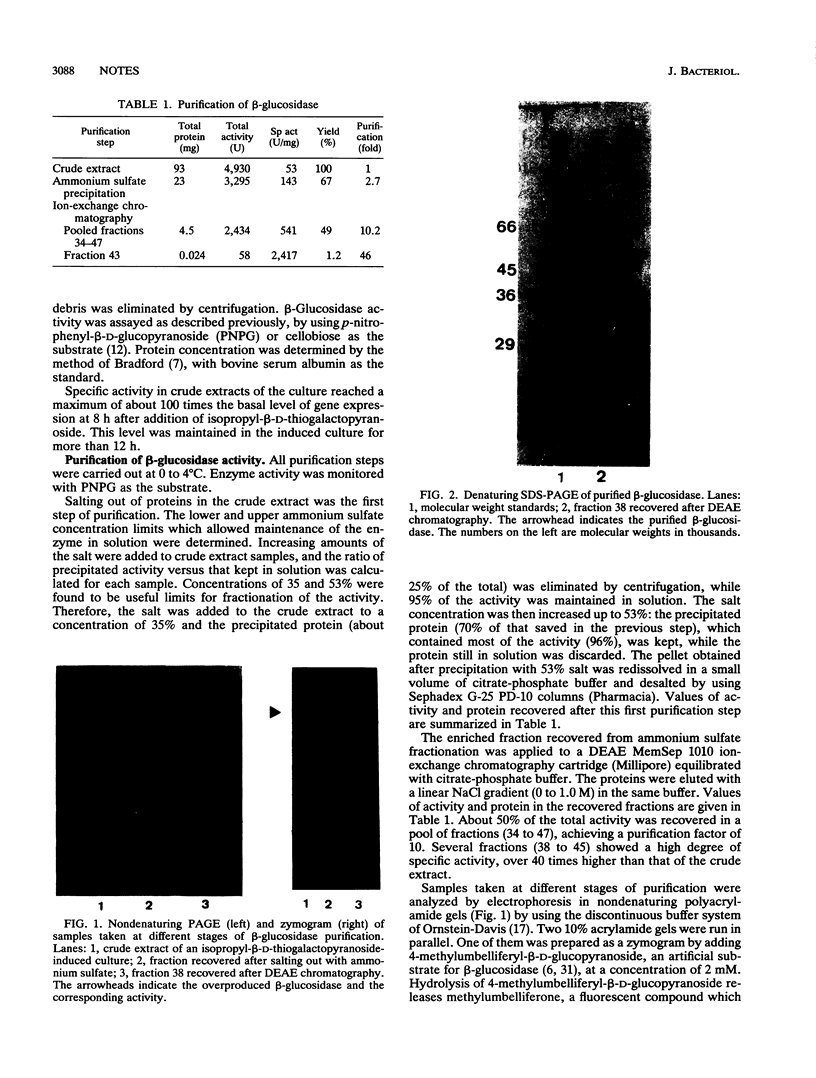
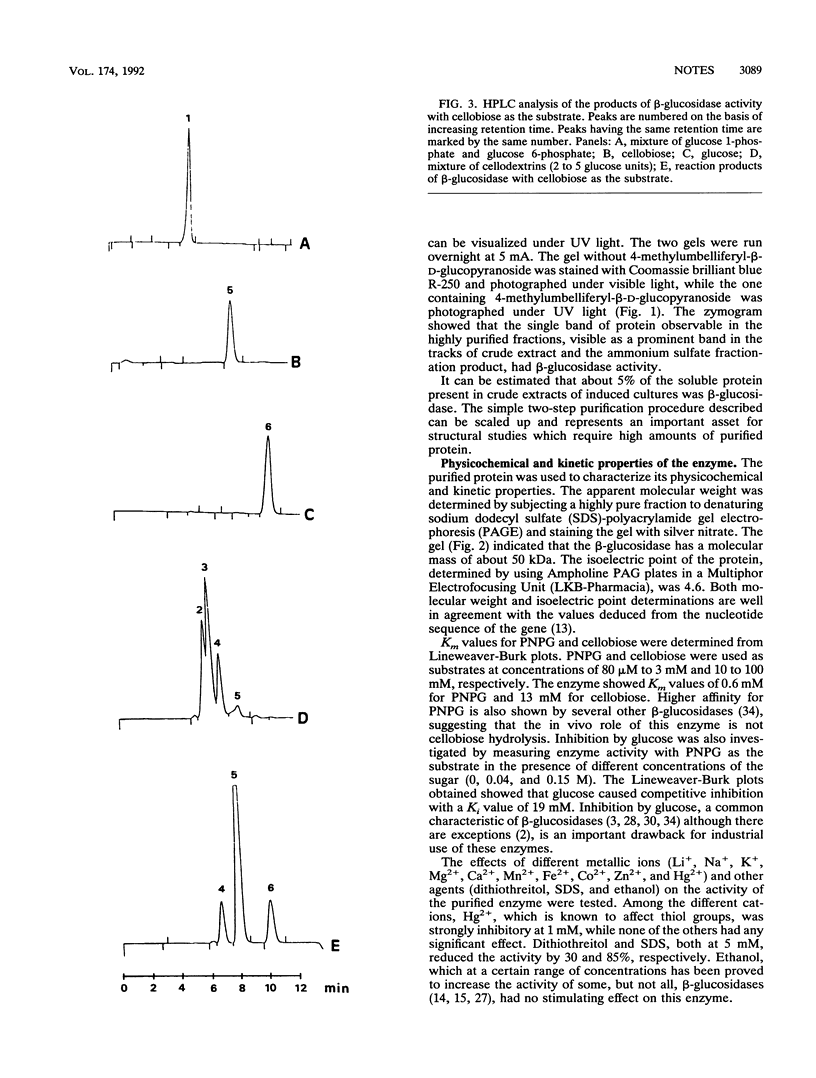
The beta-glucosidase encoded by the bglA gene from Bacillus polymyxa was overproduced in Escherichia coli by using a plasmid in which bglA is under control of the lacI promoter. Induction with isopropyl-beta-D-thiogalactopyranoside allowed an increase in the specific activity of the enzyme of about 100 times the basal level of gene expression. The enzyme was purified by a two-step procedure involving salting out with ammonium sulfate and ion-exchange chromatography with DEAE-cellulose. Fractions of beta-glucosidase activity recovered by this procedure, after electrophoresis in an acrylamide gel and staining with silver nitrate, yielded a single band of protein. This band was shown by a zymogram to correspond to beta-glucosidase activity. The purified protein showed an apparent molecular mass of 50 kDa and an isoelectric point of 4.6, values in agreement with those expected from the nucleotide sequence of the gene. Km values of the enzyme, with either cellobiose or p-nitrophenyl-beta-D-glucoside as the substrate, were determined. It was shown that the enzyme is competitively inhibited by glucose. The effects of different metallic ions and other agents were studied. Hg2+ was strongly inhibitory, while none of the other cations tested had any significant effect. Ethanol did not show the stimulating effect observed with other beta-glucosidases. The mechanism of enzyme action was investigated. High-pressure liquid chromatography analysis with cellobiose as the substrate confirmed previous data revealing the formation of two products, glucose and another, unidentified, compound. Results presented here indicate that this compound is cellotriose formed by transglycosylation.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adam A. C., Polaina J. Construction of a Saccharomyces cerevisiae strain able to ferment cellobiose. Curr Genet. 1991 Jul;20(1-2):5–8. doi: 10.1007/BF00312758. [DOI] [PubMed] [Google Scholar]
- Alexander J. K. Purification and specificity of cellobiose phosphorylase from Clostridium thermocellum. J Biol Chem. 1968 Jun 10;243(11):2899–2904. [PubMed] [Google Scholar]
- BURSTEIN C., COHN M., KEPES A., MONOD J. R OLE DU LACTOSE ET DE SES PRODUITS M'ETABOLIQUES DANS L'INDUCTION DE L'OP'ERON LACTOSE CHEZ ESCHERICHIA COLI. Biochim Biophys Acta. 1965 Apr 19;95:634–639. [PubMed] [Google Scholar]
- Barnett C. C., Berka R. M., Fowler T. Cloning and amplification of the gene encoding an extracellular beta-glucosidase from Trichoderma reesei: evidence for improved rates of saccharification of cellulosic substrates. Biotechnology (N Y) 1991 Jun;9(6):562–567. doi: 10.1038/nbt0691-562. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Desrochers M., Jurasek L., Paice M. G. High Production of beta-Glucosidase in Schizophyllum commune: Isolation of the Enzyme and Effect of the Culture Filtrate on Cellulose Hydrolysis. Appl Environ Microbiol. 1981 Jan;41(1):222–228. doi: 10.1128/aem.41.1.222-228.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estruch J. J., Chriqui D., Grossmann K., Schell J., Spena A. The plant oncogene rolC is responsible for the release of cytokinins from glucoside conjugates. EMBO J. 1991 Oct;10(10):2889–2895. doi: 10.1002/j.1460-2075.1991.tb07838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estruch J. J., Schell J., Spena A. The protein encoded by the rolB plant oncogene hydrolyses indole glucosides. EMBO J. 1991 Nov;10(11):3125–3128. doi: 10.1002/j.1460-2075.1991.tb04873.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Candelas L., Aristoy M. C., Polaina J., Flors A. Cloning and characterization of two genes from Bacillus polymyxa expressing beta-glucosidase activity in Escherichia coli. Appl Environ Microbiol. 1989 Dec;55(12):3173–3177. doi: 10.1128/aem.55.12.3173-3177.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Candelas L., Ramón D., Polaina J. Sequences and homology analysis of two genes encoding beta-glucosidases from Bacillus polymyxa. Gene. 1990 Oct 30;95(1):31–38. doi: 10.1016/0378-1119(90)90410-s. [DOI] [PubMed] [Google Scholar]
- Gopalan V., Glew R. H., Libell D. P., DePetro J. J. The dual effects of alcohols on the kinetic properties of guinea pig liver cytosolic beta-glucosidase. J Biol Chem. 1989 Sep 15;264(26):15418–15422. [PubMed] [Google Scholar]
- Grogan Dennis W. Evidence that beta-Galactosidase of Sulfolobus solfataricus Is Only One of Several Activities of a Thermostable beta-d-Glycosidase. Appl Environ Microbiol. 1991 Jun;57(6):1644–1649. doi: 10.1128/aem.57.6.1644-1649.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HULCHER F. H., KING K. W. Disaccharide preference of an aerobic cellulolytic bacterium, Cellvibrio gilvus n. sp. J Bacteriol. 1958 Dec;76(6):565–570. doi: 10.1128/jb.76.6.565-570.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüttermann A., Volger C. Cellobiose phosphorylase in Fomes annosus. Nat New Biol. 1973 Sep 12;245(141):64–64. doi: 10.1038/newbio245064a0. [DOI] [PubMed] [Google Scholar]
- Jobe A., Bourgeois S. lac Repressor-operator interaction. VI. The natural inducer of the lac operon. J Mol Biol. 1972 Aug 28;69(3):397–408. doi: 10.1016/0022-2836(72)90253-7. [DOI] [PubMed] [Google Scholar]
- Kadam S. K., Demain A. L. Addition of cloned beta-glucosidase enhances the degradation of crystalline cellulose by the Clostridium thermocellum cellulose complex. Biochem Biophys Res Commun. 1989 Jun 15;161(2):706–711. doi: 10.1016/0006-291x(89)92657-0. [DOI] [PubMed] [Google Scholar]
- MUELLER-HILL B., RICKENBERG H. V., WALLENFELS K. SPECIFICITY OF THE INDUCTION OF THE ENZYMES OF THE LAC OPERON IN ESCHERICHIA COLI. J Mol Biol. 1964 Nov;10:303–318. doi: 10.1016/s0022-2836(64)80049-8. [DOI] [PubMed] [Google Scholar]
- Ng T. K., Zeikus J. G. Differential metabolism of cellobiose and glucose by Clostridium thermocellum and Clostridium thermohydrosulfuricum. J Bacteriol. 1982 Jun;150(3):1391–1399. doi: 10.1128/jb.150.3.1391-1399.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer R. E., Anderson R. L. Cellobiose metabolism in Aerobacter aerogenes. II. Phosphorylation of cellobiose with adenosine 5'-triphosphate by a -glucoside kinase. J Biol Chem. 1972 Jun 10;247(11):3415–3419. [PubMed] [Google Scholar]
- Plant A. R., Oliver J. E., Patchett M. L., Daniel R. M., Morgan H. W. Stability and substrate specificity of a beta-glucosidase from the thermophilic bacterium Tp8 cloned into Escherichia coli. Arch Biochem Biophys. 1988 Apr;262(1):181–188. doi: 10.1016/0003-9861(88)90180-4. [DOI] [PubMed] [Google Scholar]
- Shewale J. G. Beta-Glucosidase: its role in cellulase synthesis and hydrolysis of cellulose. Int J Biochem. 1982;14(6):435–443. doi: 10.1016/0020-711x(82)90109-4. [DOI] [PubMed] [Google Scholar]
- Sternberg D., Vijayakumar P., Reese E. T. beta-Glucosidase: microbial production and effect on enzymatic hydrolysis of cellulose. Can J Microbiol. 1977 Feb;23(2):139–147. doi: 10.1139/m77-020. [DOI] [PubMed] [Google Scholar]