Abstract
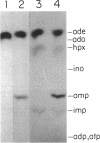
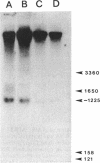
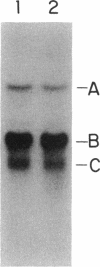
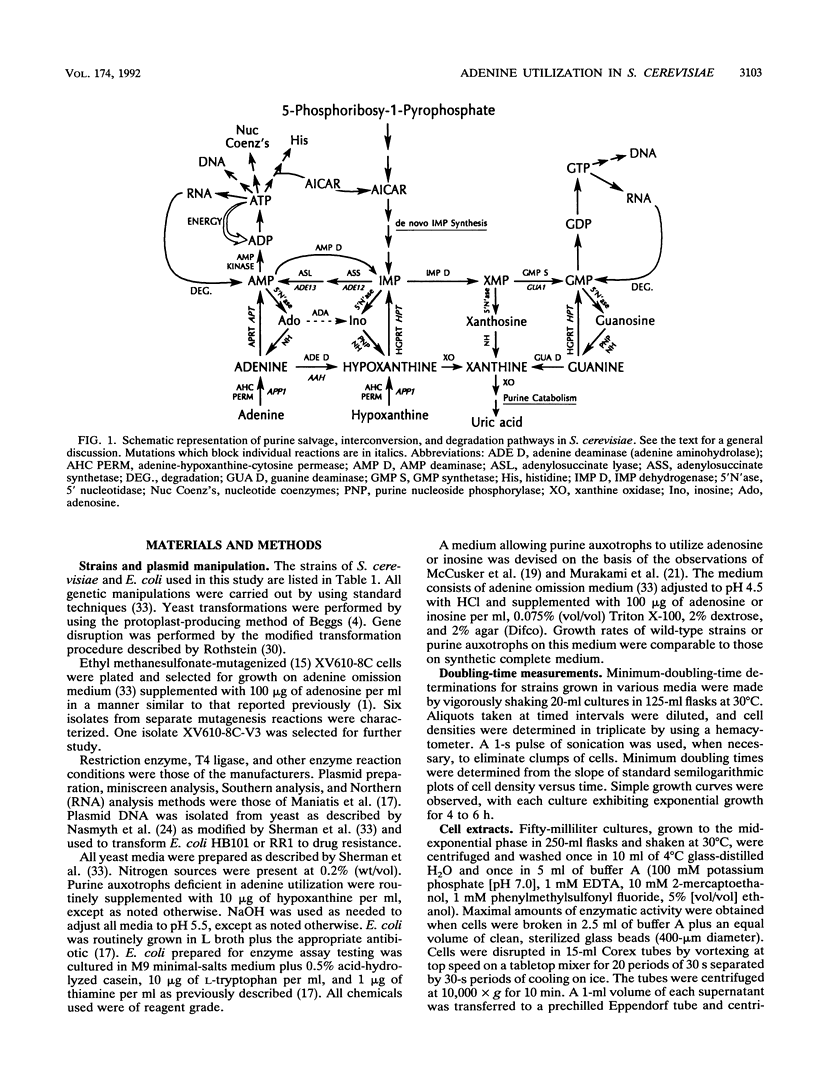
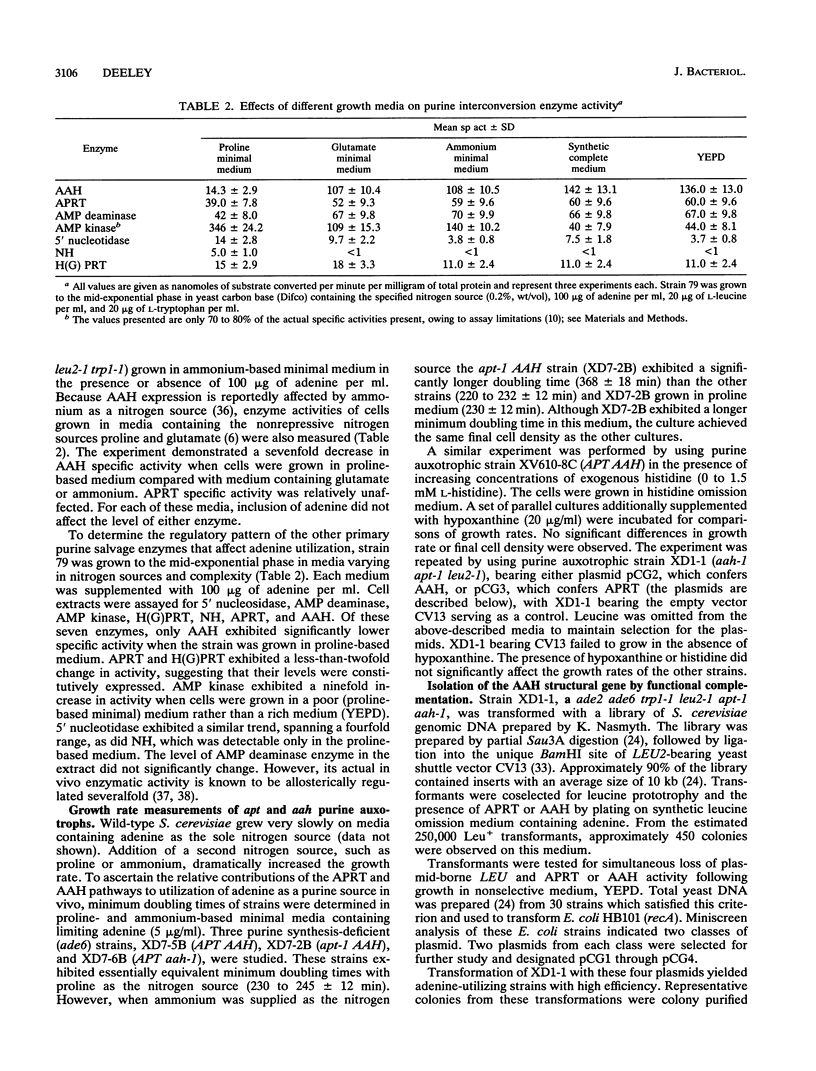
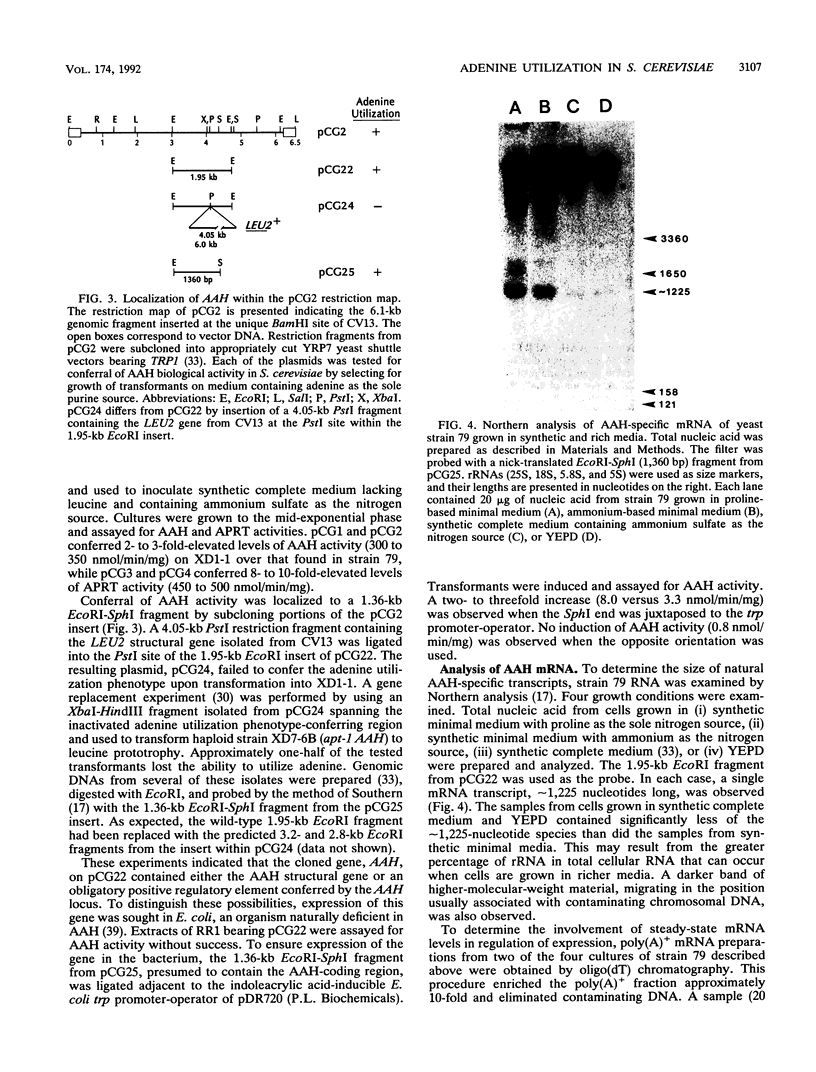
Compared with other purine salvage and nitrogen catabolism enzymatic activities, adenine deaminase (adenine aminohydrolase [AAH]; EC 3.5.4.2) activity in Saccharomyces cerevisiae is uniquely regulated. AAH specific activity is not induced by adenine and is reduced sevenfold when cells are cultivated in medium containing proline in place of ammonium as the sole nitrogen source. Exogenous adenine enters metabolic pathways primarily via the function of either AAH or adenine phosphoribosyltransferase (APRT; EC 2.4.2.7). Exogenous adenosine cannot normally be utilized as a purine source. Strains efficiently utilized adenosine or inosine when grown in pH 4.5 medium containing Triton X-100. A recessive mutation permitting utilization of adenosine or inosine in standard media was isolated. In both situations, growth of purine auxotrophs required either AAH or APRT activity. With medium containing either ammonium or proline as a nitrogen source, minimum doubling times of purine auxotrophs deficient in either APRT or AAH were measured. In proline-based medium, AAH and APRT permitted equal utilization of exogenous adenine. In ammonium-based medium, the absence of APRT increased the minimum doubling time by 50%. Similar experiments using sufficient exogenous histidine to feedback inhibit histidine biosynthesis failed to affect the growth rates of adenine auxotrophs blocked in AAH or APRT, indicating that the histidine-biosynthetic pathway does not play a significant role in adenine utilization. The gene that encodes AAH in S. cerevisiae was isolated by complementation using yeast strain XD1-1, which is deficient in AAH, APRT, and purine synthesis. A 1.36-kb EcoRI-SphI fragment was demonstrated to contain the structural gene for AAH by expressing this DNA in Escherichia coli under control of the trp promoter-operator. Northern (RNA) studies using the AAH-, APRT-, and CDC3-coding regions indicated that AAH regulation was not mediated at the level of transcription or mRNA degradation.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Armitt S., Woods R. A. Purine-excreting mutants of Saccharomyces cerevisiae. I. Isolation and genetic analysis. Genet Res. 1970 Feb;15(1):7–17. doi: 10.1017/s0016672300001324. [DOI] [PubMed] [Google Scholar]
- Ball W. J., Jr, Atkinson D. E. Adenylate energy charge in Saccharomyces cerevisiae during starvation. J Bacteriol. 1975 Mar;121(3):975–982. doi: 10.1128/jb.121.3.975-982.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beggs J. D. Transformation of yeast by a replicating hybrid plasmid. Nature. 1978 Sep 14;275(5676):104–109. doi: 10.1038/275104a0. [DOI] [PubMed] [Google Scholar]
- Burridge P. W., Woods R. A., Henderson J. F. Purine metabolism in Saccharomyces cerevisiae. Can J Biochem. 1977 Sep;55(9):935–941. doi: 10.1139/o77-140. [DOI] [PubMed] [Google Scholar]
- Gardner W. J., Woods R. A. Isolation and characterisation of guanine auxotrophs in Saccharomyces cerevisiae. Can J Microbiol. 1979 Mar;25(3):380–389. doi: 10.1139/m79-059. [DOI] [PubMed] [Google Scholar]
- HEPPEL L. A., HILMOE R. J. [Phosphorolysis and hydrolysis of purine ribosides by enzymes from yeast]. J Biol Chem. 1952 Oct;198(2):683–694. [PubMed] [Google Scholar]
- Ito Y., Tomasselli A. G., Noda L. H. ATP:AMP phosphotransferase from baker's yeast. Purification and properties. Eur J Biochem. 1980 Mar;105(1):85–92. doi: 10.1111/j.1432-1033.1980.tb04477.x. [DOI] [PubMed] [Google Scholar]
- Jakubowski H., Goldman E. Evidence for cooperation between cells during sporulation of the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1988 Dec;8(12):5166–5178. doi: 10.1128/mcb.8.12.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jochimsen B., Nygaard P., Vestergaard T. Location on the chromosome of Escherichia coli of genes governing purine metabolism. Adenosine deaminase (add), guanosine kinase (gsk) and hypoxanthine phosphoribosyltransferase (hpt). Mol Gen Genet. 1975 Dec 30;143(1):85–91. doi: 10.1007/BF00269424. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laten H. M., Valentine P. J., van Kast C. A. Adenosine accumulation in Saccharomyces cerevisiae cultured in medium containing low levels of adenine. J Bacteriol. 1986 Jun;166(3):763–768. doi: 10.1128/jb.166.3.763-768.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung H. B., Schramm V. L. A mutant AMP nucleosidase. Purification, properties, and in vivo turnover of the protein. J Biol Chem. 1981 Dec 25;256(24):12823–12829. [PubMed] [Google Scholar]
- Lindegren G., Hwang Y. L., Oshima Y., Lindegren C. C. Genetical mutants induced by ethyl methanesulfonate in Saccharomyces. Can J Genet Cytol. 1965 Sep;7(3):491–499. doi: 10.1139/g65-064. [DOI] [PubMed] [Google Scholar]
- Marmocchi F., Lupidi G., Venardi G., Riva F. Adenosine deaminase from Saccharomyces cerevisiae: purification and characterization. Biochem Int. 1987 Mar;14(3):569–580. [PubMed] [Google Scholar]
- McCusker J. H., Haber J. E. Efficient sporulation of yeast in media buffered near pH6. J Bacteriol. 1977 Oct;132(1):180–185. doi: 10.1128/jb.132.1.180-185.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami K., Nagura H., Yoshino M. Permeabilization of yeast cells: application to study on the regulation of AMP deaminase activity in situ. Anal Biochem. 1980 Jul 1;105(2):407–413. doi: 10.1016/0003-2697(80)90479-0. [DOI] [PubMed] [Google Scholar]
- Murray A. W., Elliott D. C., Atkinson M. R. Nucleotide biosynthesis from preformed purines in mammalian cells: regulatory mechanisms and biological significance. Prog Nucleic Acid Res Mol Biol. 1970;10:87–119. doi: 10.1016/s0079-6603(08)60562-0. [DOI] [PubMed] [Google Scholar]
- Murray A. W. The biological significance of purine salvage. Annu Rev Biochem. 1971;40:811–826. doi: 10.1146/annurev.bi.40.070171.004115. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. A., Reed S. I. Isolation of genes by complementation in yeast: molecular cloning of a cell-cycle gene. Proc Natl Acad Sci U S A. 1980 Apr;77(4):2119–2123. doi: 10.1073/pnas.77.4.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochs U. H., Chen S. H., Ochs H. D., Osborne W. R., Scott C. R. Purine nucleoside phosphorylase deficiency: a molecular model for selective loss of T cell function. J Immunol. 1979 Jun;122(6):2424–2429. [PubMed] [Google Scholar]
- POMPER S. Purine-requiring and pyrimidine-requiring mutants of Saccharomyces cerevisiae. J Bacteriol. 1952 Jun;63(6):707–713. doi: 10.1128/jb.63.6.707-713.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak A., Grenson M. Evidence for a common transport system for cytosine, adenine and hypoxanthine in Saccharomyces cerevisiae and Candida albicans. Eur J Biochem. 1973 Jan 15;32(2):276–282. doi: 10.1111/j.1432-1033.1973.tb02608.x. [DOI] [PubMed] [Google Scholar]
- ROUSH A. H., SAEED M. Adenine metabolism in Saccharomyces cerevisiae adenase from bakers' yeast. Biochem Biophys Res Commun. 1960 Jan;2:43–47. doi: 10.1016/0006-291x(60)90262-x. [DOI] [PubMed] [Google Scholar]
- Rasse-Messenguy F., Fink G. R. Feedback-resistant mutants of histidine biosynthesis in yeast. Basic Life Sci. 1973;2:85–95. doi: 10.1007/978-1-4684-2880-3_7. [DOI] [PubMed] [Google Scholar]
- Remy C. N., Love S. H. Induction of adenosine deaminase in Escherichia coli. J Bacteriol. 1968 Jul;96(1):76–85. doi: 10.1128/jb.96.1.76-85.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Schmidt R., Wiegand H., Reichert U. Purification and characterization of the hypoxanthine-guanine phosphoribosyltransferase from Saccharomyces cerevisiae. Eur J Biochem. 1979 Jan 15;93(2):355–361. doi: 10.1111/j.1432-1033.1979.tb12830.x. [DOI] [PubMed] [Google Scholar]
- Vogels G. D., Van der Drift C. Degradation of purines and pyrimidines by microorganisms. Bacteriol Rev. 1976 Jun;40(2):403–468. doi: 10.1128/br.40.2.403-468.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods R. A., Roberts D. G., Friedman T., Jolly D., Filpula D. Hypoxanthine: guanine phosphoribosyltransferase mutants in Saccharomyces cerevisiae. Mol Gen Genet. 1983;191(3):407–412. doi: 10.1007/BF00425755. [DOI] [PubMed] [Google Scholar]
- Woods R. A., Roberts D. G., Stein D. S., Filpula D. Adenine phosphoribosyltransferase mutants in Saccharomyces cerevisiae. J Gen Microbiol. 1984 Oct;130(10):2629–2637. doi: 10.1099/00221287-130-10-2629. [DOI] [PubMed] [Google Scholar]
- Yoshino M., Murakami K. AMP deaminase reaction as a control system of glycolysis in yeast. Role of ammonium ion in the interaction of phosphofructokinase and pyruvate kinase activity with the adenylate energy charge. J Biol Chem. 1985 Apr 25;260(8):4729–4732. [PubMed] [Google Scholar]
- Yoshino M., Murakami K., Tsushima K. AMP deaminase from baker's yeast. Purification and some regulatory properties. Biochim Biophys Acta. 1979 Sep 12;570(1):157–166. doi: 10.1016/0005-2744(79)90210-9. [DOI] [PubMed] [Google Scholar]