Abstract
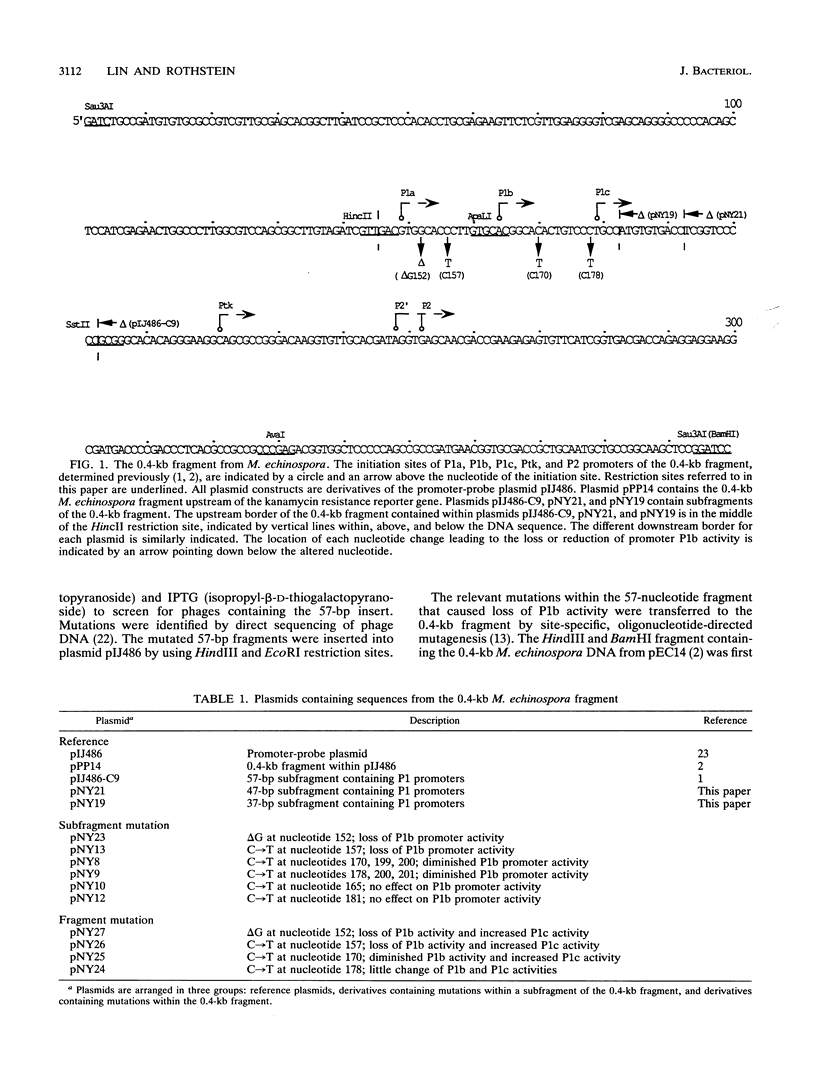
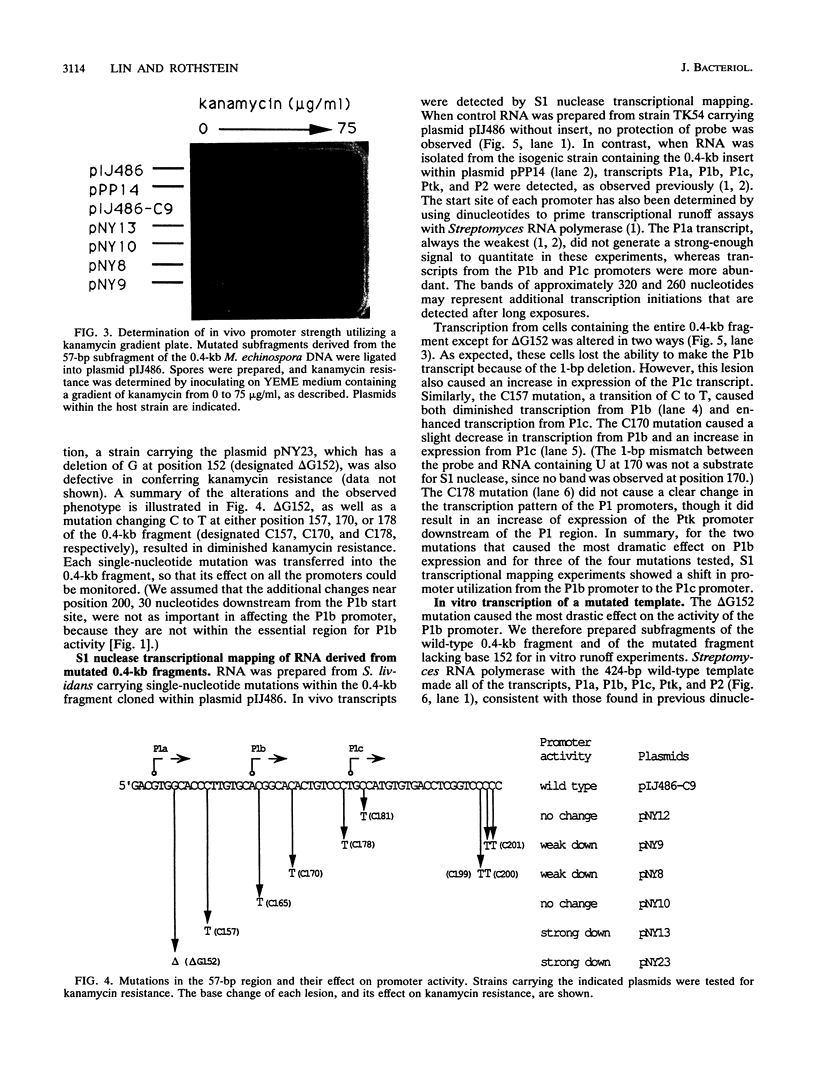
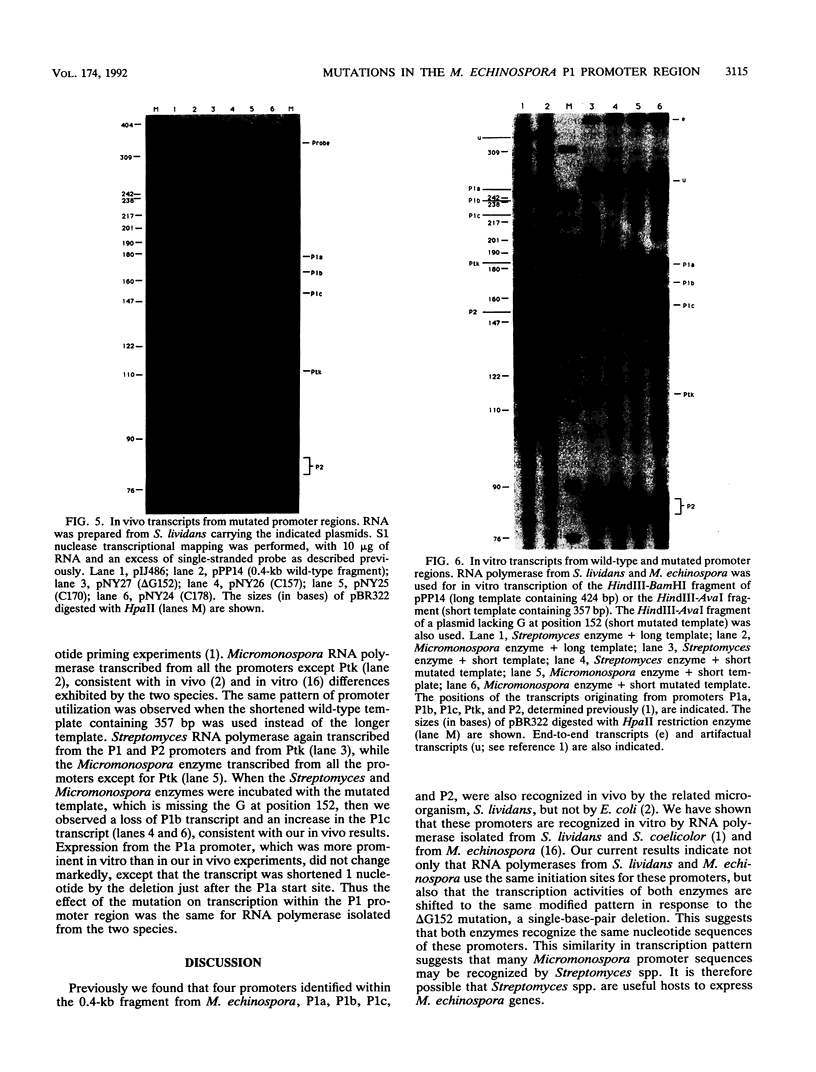
We demonstrated previously that the promoters P1a, P1b, and P1c are very closely spaced and are coordinately turned on during stationary phase in Micromonospora echinospora. To determine the nucleotides important for promoter recognition, we characterized mutations that were defective in transcription from the P1b start site, using Streptomyces lividans as the host. Two mutations upstream of the start site resulted in a drastic loss of promoter activity, while two mutations downstream of the start site resulted in a moderate loss of activity. These mutations suggest an unexpected relationship between promoters P1b and P1c. Three of the mutations that caused diminished transcription from promoter P1b simultaneously resulted in an increase in transcription from the P1c promoter initiation site located 15 bp downstream. Despite the proximity and the coordinate regulation of promoters P1b and P1c, they are in competition as transcriptional start sites.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baum E. Z., Buttner M. J., Lin L. S., Rothstein D. M. Transcription from the P1 promoters of Micromonospora echinospora in the absence of native upstream DNA sequences. J Bacteriol. 1989 Dec;171(12):6503–6510. doi: 10.1128/jb.171.12.6503-6510.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum E. Z., Love S. F., Rothstein D. M. Temporally regulated tandem promoters in Micromonospora echinospora. J Bacteriol. 1988 Jan;170(1):71–77. doi: 10.1128/jb.170.1.71-77.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogenhagen D. F., Sakonju S., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: II. The 3' border of the region. Cell. 1980 Jan;19(1):27–35. doi: 10.1016/0092-8674(80)90385-2. [DOI] [PubMed] [Google Scholar]
- Budowsky E. I. The mechanism of the mutagenic action of hydroxylamines. Prog Nucleic Acid Res Mol Biol. 1976;16:125–188. doi: 10.1016/s0079-6603(08)60757-6. [DOI] [PubMed] [Google Scholar]
- Buttner M J, Fearnley I M, Bibb M J. The agarase gene (dagA) of Streptomyces coelicolor A3(2): nucleotide sequence and transcriptional analysis. Mol Gen Genet. 1987 Aug;209(1):101–109. doi: 10.1007/BF00329843. [DOI] [PubMed] [Google Scholar]
- Buttner M. J., Brown N. L. RNA polymerase-DNA interactions in Streptomyces. In vitro studies of a S. lividans plasmid promoter with S. coelicolor RNA polymerase. J Mol Biol. 1985 Sep 5;185(1):177–188. doi: 10.1016/0022-2836(85)90189-5. [DOI] [PubMed] [Google Scholar]
- Buttner M. J., Chater K. F., Bibb M. J. Cloning, disruption, and transcriptional analysis of three RNA polymerase sigma factor genes of Streptomyces coelicolor A3(2). J Bacteriol. 1990 Jun;172(6):3367–3378. doi: 10.1128/jb.172.6.3367-3378.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttner M. J. RNA polymerase heterogeneity in Streptomyces coelicolor A3(2). Mol Microbiol. 1989 Nov;3(11):1653–1659. doi: 10.1111/j.1365-2958.1989.tb00151.x. [DOI] [PubMed] [Google Scholar]
- Buttner M. J., Smith A. M., Bibb M. J. At least three different RNA polymerase holoenzymes direct transcription of the agarase gene (dagA) of Streptomyces coelicolor A3(2). Cell. 1988 Feb 26;52(4):599–607. doi: 10.1016/0092-8674(88)90472-2. [DOI] [PubMed] [Google Scholar]
- Chater K. F., Bruton C. J., Plaskitt K. A., Buttner M. J., Méndez C., Helmann J. D. The developmental fate of S. coelicolor hyphae depends upon a gene product homologous with the motility sigma factor of B. subtilis. Cell. 1989 Oct 6;59(1):133–143. doi: 10.1016/0092-8674(89)90876-3. [DOI] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983 Jun 5;166(4):477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lee M. D., Manning J. K., Williams D. R., Kuck N. A., Testa R. T., Borders D. B. Calicheamicins, a novel family of antitumor antibiotics. 3. Isolation, purification and characterization of calicheamicins beta 1Br, gamma 1Br, alpha 2I, alpha 3I, beta 1I, gamma 1I and delta 1I. J Antibiot (Tokyo) 1989 Jul;42(7):1070–1087. doi: 10.7164/antibiotics.42.1070. [DOI] [PubMed] [Google Scholar]
- Lin L. S., Kim Y. J., Meyer R. J. The 20 bp, directly repeated DNA sequence of broad host range plasmid R1162 exerts incompatibility in vivo and inhibits R1162 DNA replication in vitro. Mol Gen Genet. 1987 Jul;208(3):390–397. doi: 10.1007/BF00328129. [DOI] [PubMed] [Google Scholar]
- Love S. F., Maiese W. M., Rothstein D. M. Conditions for protoplasting, regenerating, and transforming the calicheamicin producer, Micromonospora echinospora. Appl Environ Microbiol. 1992 Apr;58(4):1376–1378. doi: 10.1128/aem.58.4.1376-1378.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiese W. M., Lechevalier M. P., Lechevalier H. A., Korshalla J., Kuck N., Fantini A., Wildey M. J., Thomas J., Greenstein M. Calicheamicins, a novel family of antitumor antibiotics: taxonomy, fermentation and biological properties. J Antibiot (Tokyo) 1989 Apr;42(4):558–563. doi: 10.7164/antibiotics.42.558. [DOI] [PubMed] [Google Scholar]
- McClure W. R. Mechanism and control of transcription initiation in prokaryotes. Annu Rev Biochem. 1985;54:171–204. doi: 10.1146/annurev.bi.54.070185.001131. [DOI] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Rothstein D. M., Love S. F. Isolation of mutants blocked in calicheamicin biosynthesis. J Bacteriol. 1991 Dec;173(23):7716–7718. doi: 10.1128/jb.173.23.7716-7718.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. M., Janssen G. R., Kieser T., Bibb M. J., Buttner M. J., Bibb M. J. Construction and characterisation of a series of multi-copy promoter-probe plasmid vectors for Streptomyces using the aminoglycoside phosphotransferase gene from Tn5 as indicator. Mol Gen Genet. 1986 Jun;203(3):468–478. doi: 10.1007/BF00422072. [DOI] [PubMed] [Google Scholar]
- Westpheling J., Brawner M. Two transcribing activities are involved in expression of the Streptomyces galactose operon. J Bacteriol. 1989 Mar;171(3):1355–1361. doi: 10.1128/jb.171.3.1355-1361.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westpheling J., Ranes M., Losick R. RNA polymerase heterogeneity in Streptomyces coelicolor. Nature. 1985 Jan 3;313(5997):22–27. doi: 10.1038/313022a0. [DOI] [PubMed] [Google Scholar]
- Zein N., Sinha A. M., McGahren W. J., Ellestad G. A. Calicheamicin gamma 1I: an antitumor antibiotic that cleaves double-stranded DNA site specifically. Science. 1988 May 27;240(4856):1198–1201. doi: 10.1126/science.3240341. [DOI] [PubMed] [Google Scholar]