Abstract
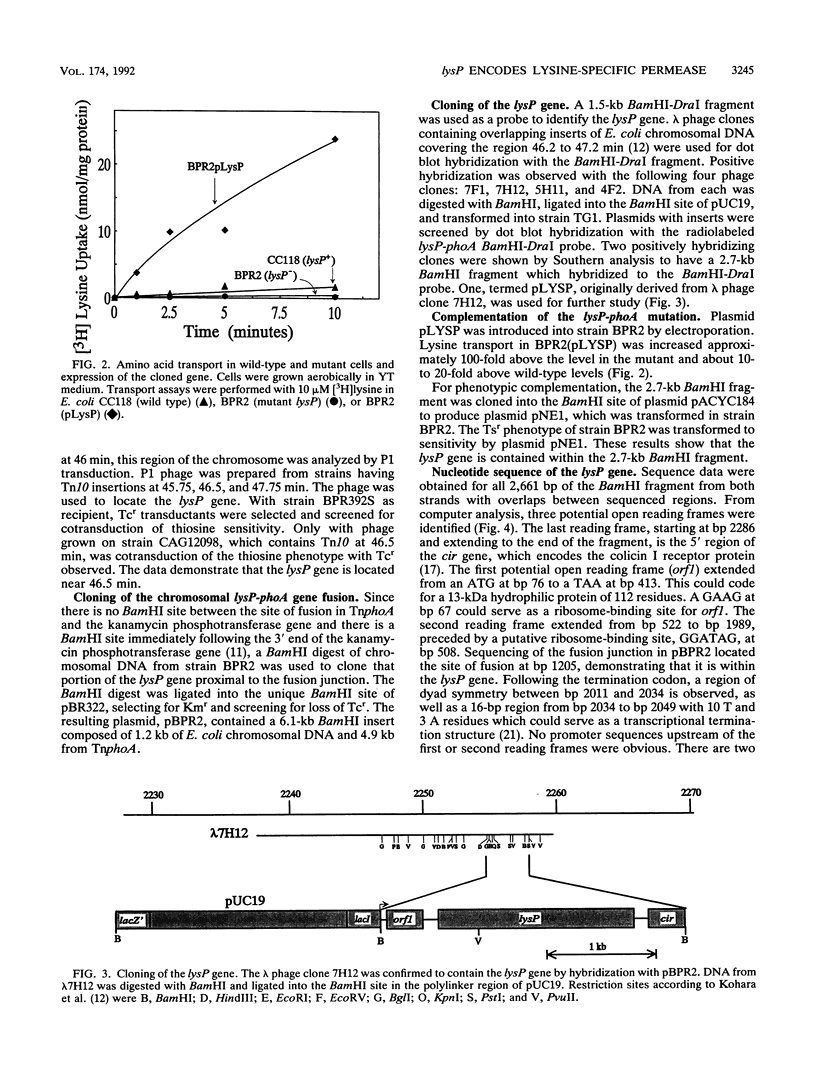
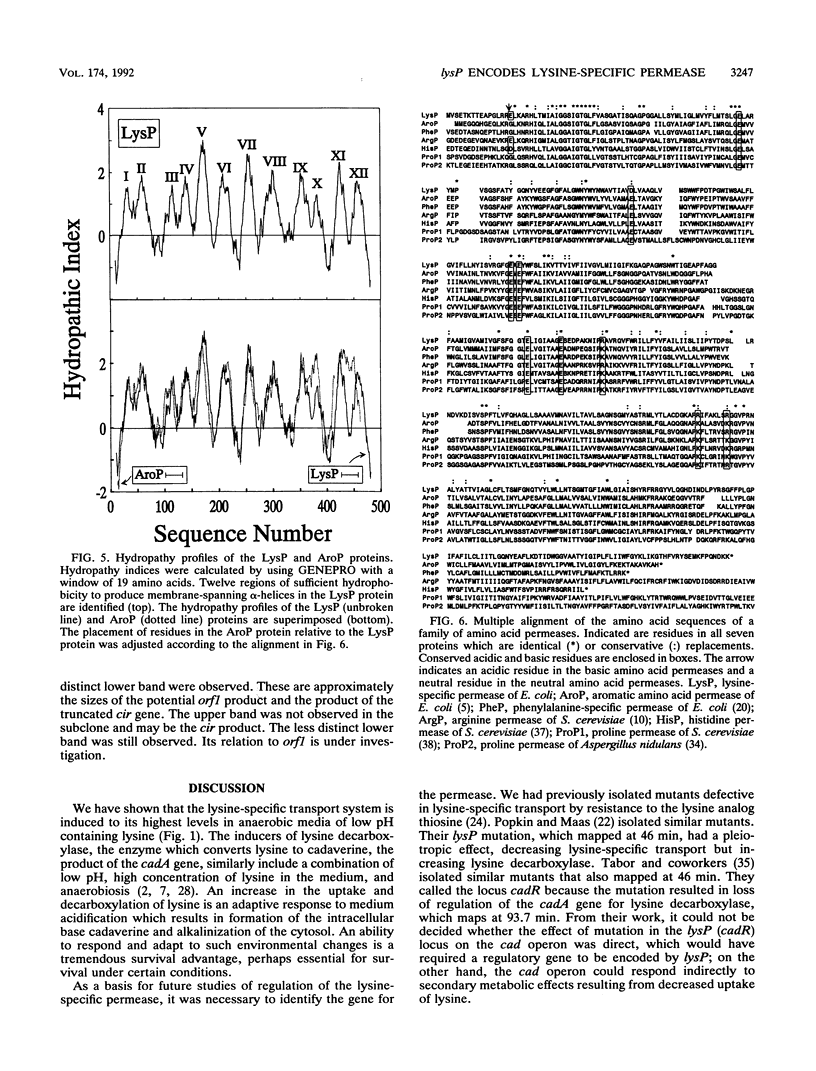
Escherichia coli transports lysine by two distinct systems, one of which is specific for lysine (LysP) and the other of which is inhibited by arginine ornithine. The activity of the lysine-specific system increases with growth in acidic medium, anaerobiosis, and high concentrations of lysine. It is inhibited by the lysine analog S-(beta-aminoethyl)-L-cysteine (thiosine). Thiosine-resistant (Tsr) mutants were isolated by using transpositional mutagenesis with TnphoA. A Tsr mutant expressing alkaline phosphatase activity in intact cells was found to lack lysine-specific transport. This lysP mutation was mapped to about 46.5 min on the E. coli chromosome. The lysP-phoA fusion was cloned and used as a probe to clone the wild-type lysP gene. The nucleotide sequence of the 2.7-kb BamHI fragment was determined. An open reading frame from nucleotides 522 to 1989 was observed. The translation product of this open reading frame is predicted to be a hydrophobic protein of 489 residues. The lysP gene product exhibits sequence similarity to a family of amino acid transport proteins found in both prokaryotes and eukaryotes, including the aromatic amino acid permease of E. coli (aroP) and the arginine permease of Saccharomyces cerevisiae (CAN1). Cells carrying a plasmid with the lysP gene exhibited a 10- to 20-fold increase in the rate of lysine uptake above wild-type levels. These results demonstrate that the lysP gene encodes the lysine-specific permease.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Auger E. A., Redding K. E., Plumb T., Childs L. C., Meng S. Y., Bennett G. N. Construction of lac fusions to the inducible arginine- and lysine decarboxylase genes of Escherichia coli K12. Mol Microbiol. 1989 May;3(5):609–620. doi: 10.1111/j.1365-2958.1989.tb00208.x. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chye M. L., Guest J. R., Pittard J. Cloning of the aroP gene and identification of its product in Escherichia coli K-12. J Bacteriol. 1986 Aug;167(2):749–753. doi: 10.1128/jb.167.2.749-753.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley C. B., Reynolds R. P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987 Mar 11;15(5):2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirshfield I. N., Zamecnik P. C. Thiosine-resistant mutants of Escherichia coli K-12 with growth-medium-dependent lysl-tRNA synthetase activity. I. Isolation and physiological characterization. Biochim Biophys Acta. 1972 Feb 15;259(3):330–343. [PubMed] [Google Scholar]
- Hoffmann W. Molecular characterization of the CAN1 locus in Saccharomyces cerevisiae. A transmembrane protein without N-terminal hydrophobic signal sequence. J Biol Chem. 1985 Sep 25;260(21):11831–11837. [PubMed] [Google Scholar]
- Jorgensen R. A., Rothstein S. J., Reznikoff W. S. A restriction enzyme cleavage map of Tn5 and location of a region encoding neomycin resistance. Mol Gen Genet. 1979;177(1):65–72. doi: 10.1007/BF00267254. [DOI] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Lüthi E., Baur H., Gamper M., Brunner F., Villeval D., Mercenier A., Haas D. The arc operon for anaerobic arginine catabolism in Pseudomonas aeruginosa contains an additional gene, arcD, encoding a membrane protein. Gene. 1990 Mar 1;87(1):37–43. doi: 10.1016/0378-1119(90)90493-b. [DOI] [PubMed] [Google Scholar]
- Manoil C., Beckwith J. TnphoA: a transposon probe for protein export signals. Proc Natl Acad Sci U S A. 1985 Dec;82(23):8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milburn M. V., Privé G. G., Milligan D. L., Scott W. G., Yeh J., Jancarik J., Koshland D. E., Jr, Kim S. H. Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science. 1991 Nov 29;254(5036):1342–1347. doi: 10.1126/science.1660187. [DOI] [PubMed] [Google Scholar]
- NOVICK R. P., MAAS W. K. Control by endogenously synthesized arginine of the formation of ornithine transcarbamylase in Escherichia coli. J Bacteriol. 1961 Feb;81:236–240. doi: 10.1128/jb.81.2.236-240.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nau C. D., Konisky J. Evolutionary relationship between the TonB-dependent outer membrane transport proteins: nucleotide and amino acid sequences of the Escherichia coli colicin I receptor gene. J Bacteriol. 1989 Feb;171(2):1041–1047. doi: 10.1128/jb.171.2.1041-1047.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peruski L. F., Jr Rapid Hfr mapping of Mu dlac fusions. Biotechniques. 1990 Jul;9(1):40–42. [PubMed] [Google Scholar]
- Pi J., Wookey P. J., Pittard A. J. Cloning and sequencing of the pheP gene, which encodes the phenylalanine-specific transport system of Escherichia coli. J Bacteriol. 1991 Jun;173(12):3622–3629. doi: 10.1128/jb.173.12.3622-3629.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- Popkin P. S., Maas W. K. Escherichia coli regulatory mutation affecting lysine transport and lysine decarboxylase. J Bacteriol. 1980 Feb;141(2):485–492. doi: 10.1128/jb.141.2.485-492.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen B. P. Basic amino acid transport in Escherichia coli. II. Purification and properties of an arginine-specific binding protein. J Biol Chem. 1973 Feb 25;248(4):1211–1218. [PubMed] [Google Scholar]
- Rosen B. P. Basic amino acid transport in Escherichia coli. J Biol Chem. 1971 Jun 10;246(11):3653–3662. [PubMed] [Google Scholar]
- Rosen B. P. Basic amino acid transport in Escherichia coli: properties of canavanine-resistant mutants. J Bacteriol. 1973 Nov;116(2):627–635. doi: 10.1128/jb.116.2.627-635.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russel M., Model P. Replacement of the fip gene of Escherichia coli by an inactive gene cloned on a plasmid. J Bacteriol. 1984 Sep;159(3):1034–1039. doi: 10.1128/jb.159.3.1034-1039.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabo D. L., Boeker E. A., Byers B., Waron H., Fischer E. H. Purification and physical properties of inducible Escherichia coli lysine decarboxylase. Biochemistry. 1974 Feb 12;13(4):662–670. doi: 10.1021/bi00701a005. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987 Jul;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saporito S. M., Cunningham R. P. Nucleotide sequence of the nfo gene of Escherichia coli K-12. J Bacteriol. 1988 Nov;170(11):5141–5145. doi: 10.1128/jb.170.11.5141-5145.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarsero J. P., Wookey P. J., Gollnick P., Yanofsky C., Pittard A. J. A new family of integral membrane proteins involved in transport of aromatic amino acids in Escherichia coli. J Bacteriol. 1991 May;173(10):3231–3234. doi: 10.1128/jb.173.10.3231-3234.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M., Baker T. A., Schnitzler G., Deischel S. M., Goel M., Dove W., Jaacks K. J., Grossman A. D., Erickson J. W., Gross C. A. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989 Mar;53(1):1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sophianopoulou V., Scazzocchio C. The proline transport protein of Aspergillus nidulans is very similar to amino acid transporters of Saccharomyces cerevisiae. Mol Microbiol. 1989 Jun;3(6):705–714. doi: 10.1111/j.1365-2958.1989.tb00219.x. [DOI] [PubMed] [Google Scholar]
- Tabor H., Hafner E. W., Tabor C. W. Construction of an Escherichia coli strain unable to synthesize putrescine, spermidine, or cadaverine: characterization of two genes controlling lysine decarboxylase. J Bacteriol. 1980 Dec;144(3):952–956. doi: 10.1128/jb.144.3.952-956.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka J., Fink G. R. The histidine permease gene (HIP1) of Saccharomyces cerevisiae. Gene. 1985;38(1-3):205–214. doi: 10.1016/0378-1119(85)90219-7. [DOI] [PubMed] [Google Scholar]
- Vandenbol M., Jauniaux J. C., Grenson M. Nucleotide sequence of the Saccharomyces cerevisiae PUT4 proline-permease-encoding gene: similarities between CAN1, HIP1 and PUT4 permeases. Gene. 1989 Nov 15;83(1):153–159. doi: 10.1016/0378-1119(89)90413-7. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]




