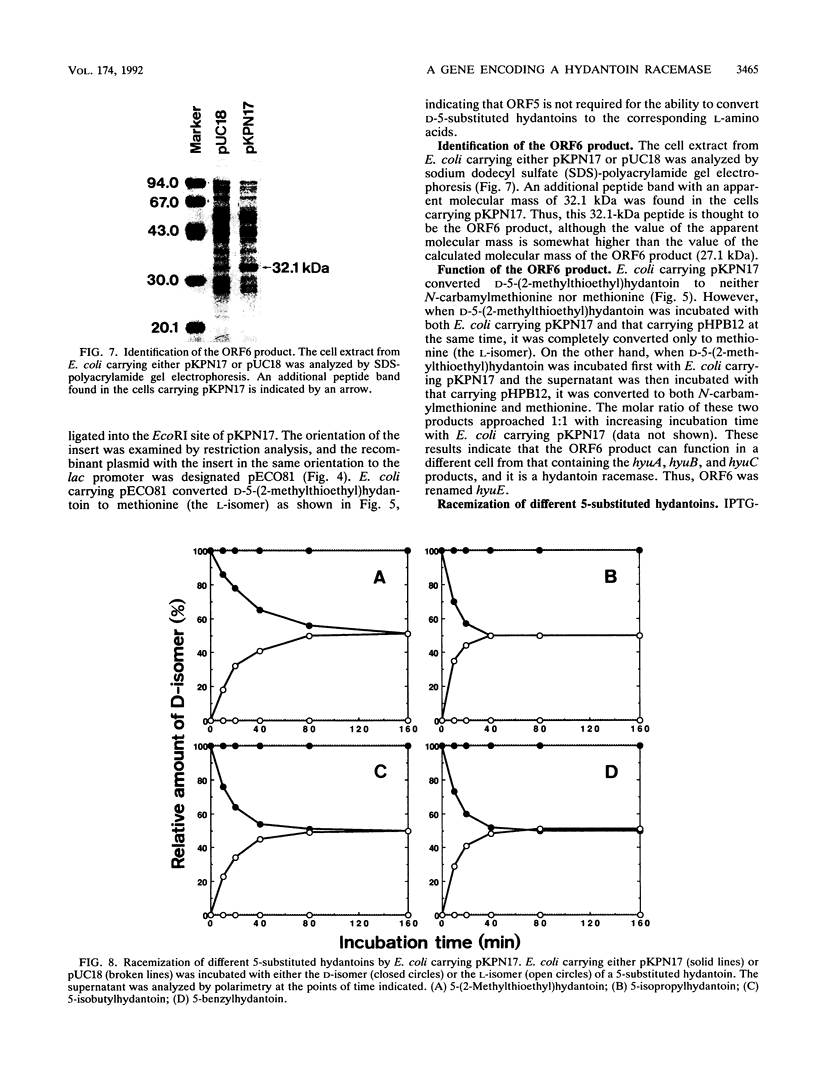
Abstract
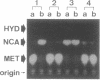
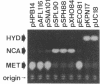
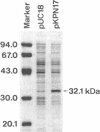
DNA fragments containing the genes involved in the conversion of 5-substituted hydantoins to their corresponding L-amino acids have been cloned from the 172-kb native plasmid (pHN671) of Pseudomonas sp. strain NS671. The largest recombinant plasmid, designated pHPB14, encoded the ability to convert D-5-substituted hydantoins to the corresponding L-amino acids, whereas the smallest one, designated pHPB12, encoded the ability to convert them to their corresponding N-carbamyl-D-amino acids. Restriction analysis suggested that the inserts of both recombinant plasmids are derived from the identical portion in pHN671 and that the insert of pHPB14, compared with that of pHPB12, has an extra 5.3 kb in length. DNA sequencing revealed that pHPB14 contains two additional complete open reading frames, designated ORF5 and hyuE. Analysis of deletion derivatives of pHPB14 indicated that hyuE is required for the ability to produce L-amino acids from the corresponding D-5-substituted hydantoins, but ORF5 is not. Cells of Escherichia coli transformed with a plasmid containing hyuE were capable of racemizing different 5-substituted hydantoins, indicating that hyuE is a gene encoding a hydantoin racemase.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- STARK G. R., SMYTH D. G. The use of cyanate for the determination of NH2-terminal residues in proteins. J Biol Chem. 1963 Jan;238:214–226. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watabe K., Ishikawa T., Mukohara Y., Nakamura H. Cloning and sequencing of the genes involved in the conversion of 5-substituted hydantoins to the corresponding L-amino acids from the native plasmid of Pseudomonas sp. strain NS671. J Bacteriol. 1992 Feb;174(3):962–969. doi: 10.1128/jb.174.3.962-969.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winegard H. M., Toennies G., Block R. J. Detection of Sulfur-containing Amino Acids on Paper Chromatograms. Science. 1948 Nov 5;108(2810):506–507. doi: 10.1126/science.108.2810.506. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]