Abstract
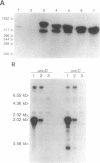
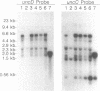
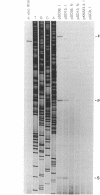
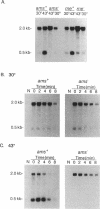
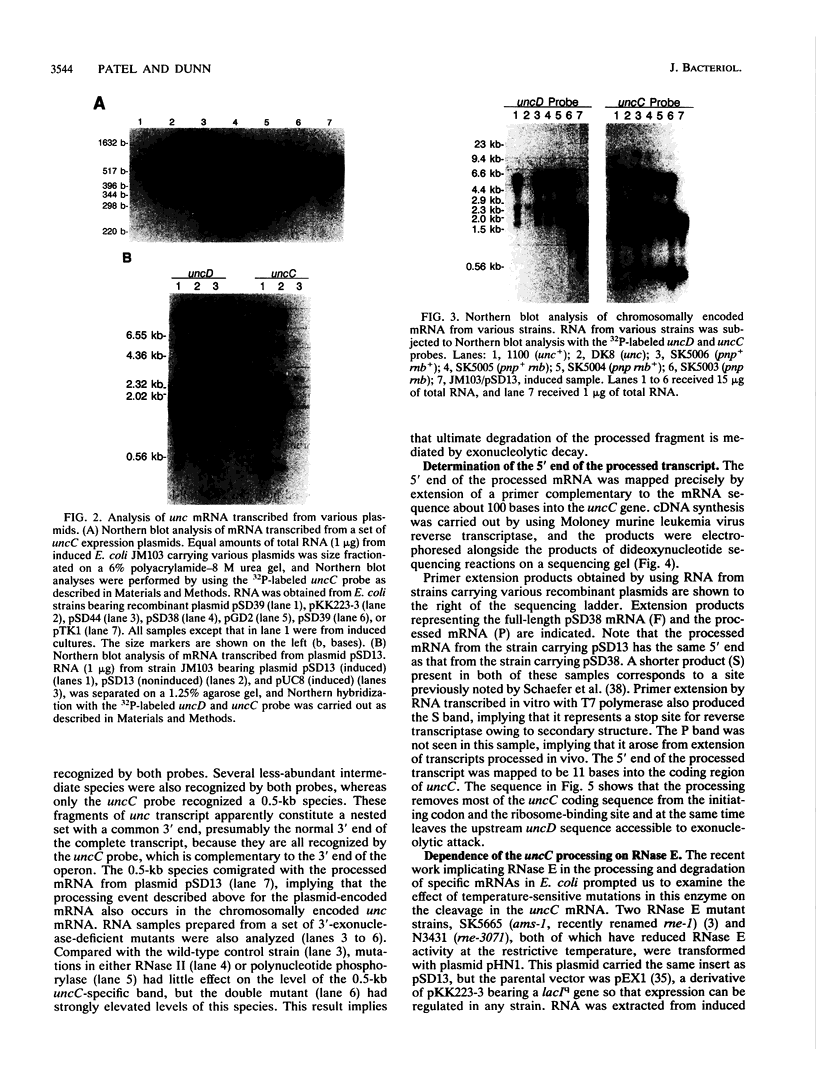
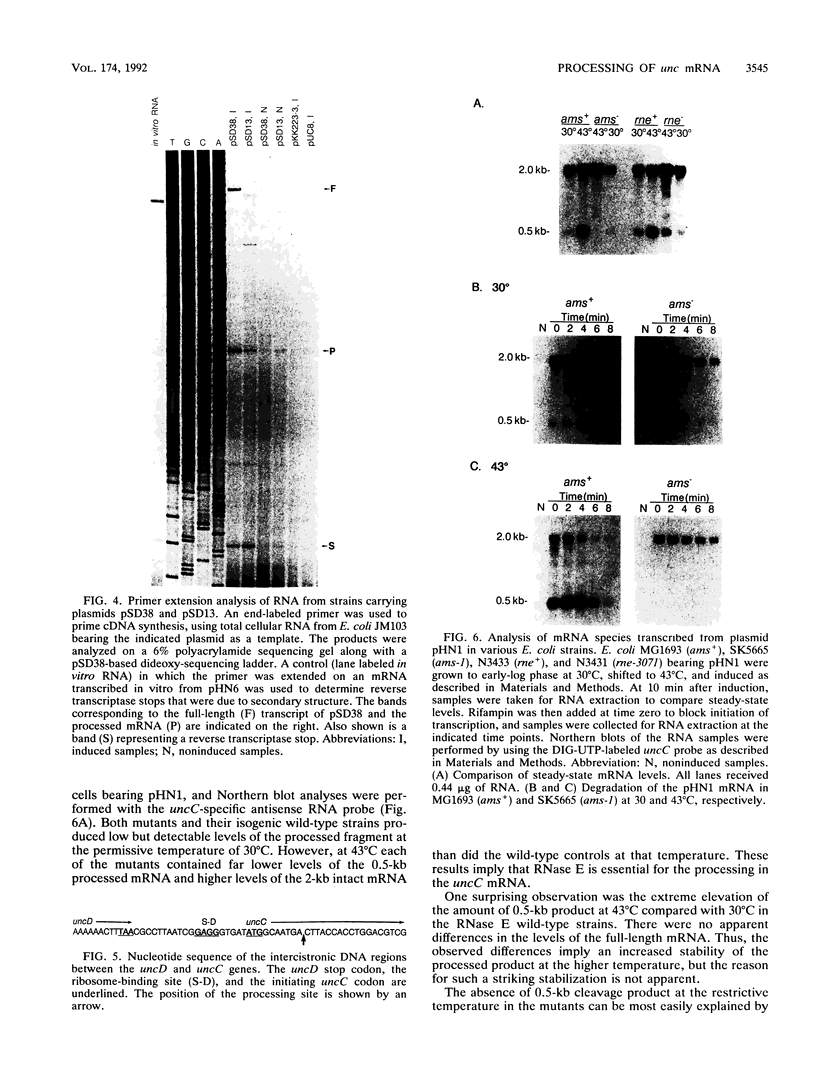
The endonucleolytic processing of the unc mRNA encoding the eight subunits of the Escherichia coli F1F0-ATPase was studied. Northern (RNA) blots of mRNA expressed from a plasmid which contained the 3'-terminal portion of the operon including the uncDC sequences revealed, in addition to the expected 2-kb mRNA, a 0.5-kb RNA species which hybridized to an uncC antisense RNA probe. An uncD antisense RNA probe hybridized to only the 2-kb mRNA, implying that the upstream 1.5-kb fragment is rapidly degraded. The 5' end of the 0.5-kb fragment was determined by primer extension analysis to be 11 bases into the coding region of the uncC gene. In RNase E-deficient strains, the amount of the 0.5-kb product was strongly reduced while the levels of the precursor uncDC transcript remained high. Similar RNase E-dependent processing was found in the chromosomally encoded unc mRNA. As this RNase E-dependent cleavage directly inactivates uncC and appears to leave uncD susceptible to degradation, it seems unlikely to play a role in differential expression of the gene products but may be an important event in unc mRNA degradation. RNase E mutants also showed altered processing of the chromosomally encoded unc mRNA in the uncB region near the 5' end. The expected full-length (7-kb) transcript was recognized when RNA from the RNase E-deficient strain was subjected to Northern blot analysis with uncB- and uncC-specific probes. RNA from strains with functional RNase E lacked the 7-kb transcript but had a 6.2-kb mRNA detectable with the uncC but not the uncB probe. RNase E is therefore implicated in multiple cleavages of the unc mRNA.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Apirion D. Isolation, genetic mapping and some characterization of a mutation in Escherichia coli that affects the processing of ribonuleic acid. Genetics. 1978 Dec;90(4):659–671. doi: 10.1093/genetics/90.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arraiano C. M., Yancey S. D., Kushner S. R. Stabilization of discrete mRNA breakdown products in ams pnp rnb multiple mutants of Escherichia coli K-12. J Bacteriol. 1988 Oct;170(10):4625–4633. doi: 10.1128/jb.170.10.4625-4633.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babitzke P., Kushner S. R. The Ams (altered mRNA stability) protein and ribonuclease E are encoded by the same structural gene of Escherichia coli. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):1–5. doi: 10.1073/pnas.88.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belasco J. G., Higgins C. F. Mechanisms of mRNA decay in bacteria: a perspective. Gene. 1988 Dec 10;72(1-2):15–23. doi: 10.1016/0378-1119(88)90123-0. [DOI] [PubMed] [Google Scholar]
- Brosius J., Holy A. Regulation of ribosomal RNA promoters with a synthetic lac operator. Proc Natl Acad Sci U S A. 1984 Nov;81(22):6929–6933. doi: 10.1073/pnas.81.22.6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusilow W. S., Klionsky D. J., Simoni R. D. Differential polypeptide synthesis of the proton-translocating ATPase of Escherichia coli. J Bacteriol. 1982 Sep;151(3):1363–1371. doi: 10.1128/jb.151.3.1363-1371.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusilow W. S., Porter A. C., Simoni R. D. Cloning and expression of uncI, the first gene of the unc operon of Escherichia coli. J Bacteriol. 1983 Sep;155(3):1265–1270. doi: 10.1128/jb.155.3.1265-1270.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Båga M., Göransson M., Normark S., Uhlin B. E. Processed mRNA with differential stability in the regulation of E. coli pilin gene expression. Cell. 1988 Jan 29;52(2):197–206. doi: 10.1016/0092-8674(88)90508-9. [DOI] [PubMed] [Google Scholar]
- Carter P. Improved oligonucleotide-directed mutagenesis using M13 vectors. Methods Enzymol. 1987;154:382–403. doi: 10.1016/0076-6879(87)54086-1. [DOI] [PubMed] [Google Scholar]
- Donovan W. P., Kushner S. R. Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc Natl Acad Sci U S A. 1986 Jan;83(1):120–124. doi: 10.1073/pnas.83.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn S. D., Dallmann H. G. An upstream uncD sequence modulates translation of Escherichia coli uncC. J Bacteriol. 1990 May;172(5):2782–2784. doi: 10.1128/jb.172.5.2782-2784.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eya S., Maeda M., Tomochika K., Kanemasa Y., Futai M. Overproduction of truncated subunit a of H+-ATPase causes growth inhibition of Escherichia coli. J Bacteriol. 1989 Dec;171(12):6853–6858. doi: 10.1128/jb.171.12.6853-6858.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster D. L., Fillingame R. H. Stoichiometry of subunits in the H+-ATPase complex of Escherichia coli. J Biol Chem. 1982 Feb 25;257(4):2009–2015. [PubMed] [Google Scholar]
- Goldblum K., Apririon D. Inactivation of the ribonucleic acid-processing enzyme ribonuclease E blocks cell division. J Bacteriol. 1981 Apr;146(1):128–132. doi: 10.1128/jb.146.1.128-132.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotta S. L., Miller O. L., Jr, French S. L. rRNA transcription rate in Escherichia coli. J Bacteriol. 1991 Oct;173(20):6647–6649. doi: 10.1128/jb.173.20.6647-6649.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross G. RNase E cleavage in the atpE leader region of atpE/interferon-beta hybrid transcripts in Escherichia coli causes enhanced rates of mRNA decay. J Biol Chem. 1991 Sep 25;266(27):17880–17884. [PubMed] [Google Scholar]
- Gunsalus R. P., Brusilow W. S., Simoni R. D. Gene order and gene-polypeptide relationships of the proton-translocating ATPase operon (unc) of Escherichia coli. Proc Natl Acad Sci U S A. 1982 Jan;79(2):320–324. doi: 10.1073/pnas.79.2.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones H. M., Brajkovich C. M., Gunsalus R. P. In vivo 5' terminus and length of the mRNA for the proton-translocating ATPase (unc) operon of Escherichia coli. J Bacteriol. 1983 Sep;155(3):1279–1287. doi: 10.1128/jb.155.3.1279-1287.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D. J., Brusilow W. S., Simoni R. D. In vivo evidence for the role of the epsilon subunit as an inhibitor of the proton-translocating ATPase of Escherichia coli. J Bacteriol. 1984 Dec;160(3):1055–1060. doi: 10.1128/jb.160.3.1055-1060.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwano M., Ono M., Endo H., Hori K., Nakamura K., Hirota Y., Ohnishi Y. Gene affecting longevity of messenger RNA: a mutant of Escherichia coli with altered mRNA stability. Mol Gen Genet. 1977 Sep 9;154(3):279–285. doi: 10.1007/BF00571283. [DOI] [PubMed] [Google Scholar]
- Mackie G. A. Stabilization of the 3' one-third of Escherichia coli ribosomal protein S20 mRNA in mutants lacking polynucleotide phosphorylase. J Bacteriol. 1989 Aug;171(8):4112–4120. doi: 10.1128/jb.171.8.4112-4120.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie G. A. Structure of the DNA distal to the gene for ribosomal protein S20 in Escherichia coli K12: presence of a strong terminator and an IS1 element. Nucleic Acids Res. 1986 Sep 11;14(17):6965–6981. doi: 10.1093/nar/14.17.6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy J. E. Expression of the unc genes in Escherichia coli. J Bioenerg Biomembr. 1988 Feb;20(1):19–39. doi: 10.1007/BF00762136. [DOI] [PubMed] [Google Scholar]
- McCarthy J. E., Gerstel B., Surin B., Wiedemann U., Ziemke P. Differential gene expression from the Escherichia coli atp operon mediated by segmental differences in mRNA stability. Mol Microbiol. 1991 Oct;5(10):2447–2458. doi: 10.1111/j.1365-2958.1991.tb02090.x. [DOI] [PubMed] [Google Scholar]
- McCarthy J. E., Schairer H. U., Sebald W. Translational initiation frequency of atp genes from Escherichia coli: identification of an intercistronic sequence that enhances translation. EMBO J. 1985 Feb;4(2):519–526. doi: 10.1002/j.1460-2075.1985.tb03659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy J. E., Schauder B., Ziemke P. Post-transcriptional control in Escherichia coli: translation and degradation of the atp operon mRNA. Gene. 1988 Dec 10;72(1-2):131–139. doi: 10.1016/0378-1119(88)90135-7. [DOI] [PubMed] [Google Scholar]
- Mead D. A., Szczesna-Skorupa E., Kemper B. Single-stranded DNA 'blue' T7 promoter plasmids: a versatile tandem promoter system for cloning and protein engineering. Protein Eng. 1986 Oct-Nov;1(1):67–74. doi: 10.1093/protein/1.1.67. [DOI] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra T. K., Apirion D. RNase E, an RNA processing enzyme from Escherichia coli. J Biol Chem. 1979 Nov 10;254(21):11154–11159. [PubMed] [Google Scholar]
- Mudd E. A., Carpousis A. J., Krisch H. M. Escherichia coli RNase E has a role in the decay of bacteriophage T4 mRNA. Genes Dev. 1990 May;4(5):873–881. doi: 10.1101/gad.4.5.873. [DOI] [PubMed] [Google Scholar]
- Mudd E. A., Krisch H. M., Higgins C. F. RNase E, an endoribonuclease, has a general role in the chemical decay of Escherichia coli mRNA: evidence that rne and ams are the same genetic locus. Mol Microbiol. 1990 Dec;4(12):2127–2135. doi: 10.1111/j.1365-2958.1990.tb00574.x. [DOI] [PubMed] [Google Scholar]
- Newbury S. F., Smith N. H., Higgins C. F. Differential mRNA stability controls relative gene expression within a polycistronic operon. Cell. 1987 Dec 24;51(6):1131–1143. doi: 10.1016/0092-8674(87)90599-x. [DOI] [PubMed] [Google Scholar]
- Nilsson P., Uhlin B. E. Differential decay of a polycistronic Escherichia coli transcript is initiated by RNaseE-dependent endonucleolytic processing. Mol Microbiol. 1991 Jul;5(7):1791–1799. doi: 10.1111/j.1365-2958.1991.tb01928.x. [DOI] [PubMed] [Google Scholar]
- Passador L., Linn T. Autogenous regulation of the RNA polymerase beta subunit of Escherichia coli occurs at the translational level in vivo. J Bacteriol. 1989 Nov;171(11):6234–6242. doi: 10.1128/jb.171.11.6234-6242.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A. M., Dallmann H. G., Skakoon E. N., Kapala T. D., Dunn S. D. The Escherichia coli unc transcription terminator enhances expression of uncC, encoding the epsilon subunit of F1-ATPase, from plasmids by stabilizing the transcript. Mol Microbiol. 1990 Nov;4(11):1941–1946. doi: 10.1111/j.1365-2958.1990.tb02043.x. [DOI] [PubMed] [Google Scholar]
- Schaefer E. M., Hartz D., Gold L., Simoni R. D. Ribosome-binding sites and RNA-processing sites in the transcript of the Escherichia coli unc operon. J Bacteriol. 1989 Jul;171(7):3901–3908. doi: 10.1128/jb.171.7.3901-3908.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taraseviciene L., Miczak A., Apirion D. The gene specifying RNase E (rne) and a gene affecting mRNA stability (ams) are the same gene. Mol Microbiol. 1991 Apr;5(4):851–855. doi: 10.1111/j.1365-2958.1991.tb00758.x. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Gay N. J. The unc operon. Nucleotide sequence, regulation and structure of ATP-synthase. Biochim Biophys Acta. 1984 Sep 6;768(2):164–200. doi: 10.1016/0304-4173(84)90003-x. [DOI] [PubMed] [Google Scholar]