Abstract
Sequence analysis of the tcmA tetracenomycin C resistance gene from Streptomyces glaucescens GLA.O (ETH 22794) identifies one large open reading frame whose deduced product has sequence similarity to the mmr methylenomycin resistance gene from Streptomyces coelicolor, the Streptomyces rimosus tet347 (otrB) tetracycline resistance gene, and the atr1 aminotriazole resistance gene from Saccharomyces cerevisiae. These genes are thought to encode proteins that act as metabolite export pumps powered by transmembrane electrochemical gradients. A divergently transcribed gene, tcmR, is located in the region upstream of tcmA. The deduced product of tcmR resembles the repressor proteins encoded by tetR regulatory genes from Escherichia coli and the actII-orf1 gene from S. coelicolor. Transcriptional analysis of tcmA and tcmR indicates that these genes have back-to-back and overlapping promoter regions.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aldea M., Claverie-Martín F., Díaz-Torres M. R., Kushner S. R. Transcript mapping using [35S]DNA probes, trichloroacetate solvent and dideoxy sequencing ladders: a rapid method for identification of transcriptional start points. Gene. 1988 May 15;65(1):101–110. doi: 10.1016/0378-1119(88)90421-0. [DOI] [PubMed] [Google Scholar]
- Ames G. F., Joshi A. K. Energy coupling in bacterial periplasmic permeases. J Bacteriol. 1990 Aug;172(8):4133–4137. doi: 10.1128/jb.172.8.4133-4137.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benveniste R., Davies J. Aminoglycoside antibiotic-inactivating enzymes in actinomycetes similar to those present in clinical isolates of antibiotic-resistant bacteria. Proc Natl Acad Sci U S A. 1973 Aug;70(8):2276–2280. doi: 10.1073/pnas.70.8.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb M. J., Findlay P. R., Johnson M. W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984 Oct;30(1-3):157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- Bormann C., Aberle K., Fiedler H. P., Schrempf H. Genetic complementation of Streptomyces tendae deficient in nikkomycin production. Appl Microbiol Biotechnol. 1990 Jan;32(4):424–430. doi: 10.1007/BF00903777. [DOI] [PubMed] [Google Scholar]
- Caballero J. L., Malpartida F., Hopwood D. A. Transcriptional organization and regulation of an antibiotic export complex in the producing Streptomyces culture. Mol Gen Genet. 1991 Sep;228(3):372–380. doi: 10.1007/BF00260629. [DOI] [PubMed] [Google Scholar]
- Cundliffe E. How antibiotic-producing organisms avoid suicide. Annu Rev Microbiol. 1989;43:207–233. doi: 10.1146/annurev.mi.43.100189.001231. [DOI] [PubMed] [Google Scholar]
- Deng Z. X., Kieser T., Hopwood D. A. Activity of a Streptomyces transcriptional terminator in Escherichia coli. Nucleic Acids Res. 1987 Mar 25;15(6):2665–2675. doi: 10.1093/nar/15.6.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert B., Beck C. F. Topology of the transposon Tn10-encoded tetracycline resistance protein within the inner membrane of Escherichia coli. J Biol Chem. 1989 Jul 15;264(20):11663–11670. [PubMed] [Google Scholar]
- Fernández-Moreno M. A., Caballero J. L., Hopwood D. A., Malpartida F. The act cluster contains regulatory and antibiotic export genes, direct targets for translational control by the bldA tRNA gene of Streptomyces. Cell. 1991 Aug 23;66(4):769–780. doi: 10.1016/0092-8674(91)90120-n. [DOI] [PubMed] [Google Scholar]
- Fisher S. H., Wray L. V., Jr Regulation of glutamine synthetase in Streptomyces coelicolor. J Bacteriol. 1989 May;171(5):2378–2383. doi: 10.1128/jb.171.5.2378-2383.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornwald J. A., Schmidt F. J., Adams C. W., Rosenberg M., Brawner M. E. Two promoters, one inducible and one constitutive, control transcription of the Streptomyces lividans galactose operon. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2130–2134. doi: 10.1073/pnas.84.8.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribskov M., Burgess R. R. Sigma factors from E. coli, B. subtilis, phage SP01, and phage T4 are homologous proteins. Nucleic Acids Res. 1986 Aug 26;14(16):6745–6763. doi: 10.1093/nar/14.16.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoile P. G., Hutchinson C. R. A bacterial analog of the mdr gene of mammalian tumor cells is present in Streptomyces peucetius, the producer of daunorubicin and doxorubicin. Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8553–8557. doi: 10.1073/pnas.88.19.8553. [DOI] [PMC free article] [PubMed] [Google Scholar]
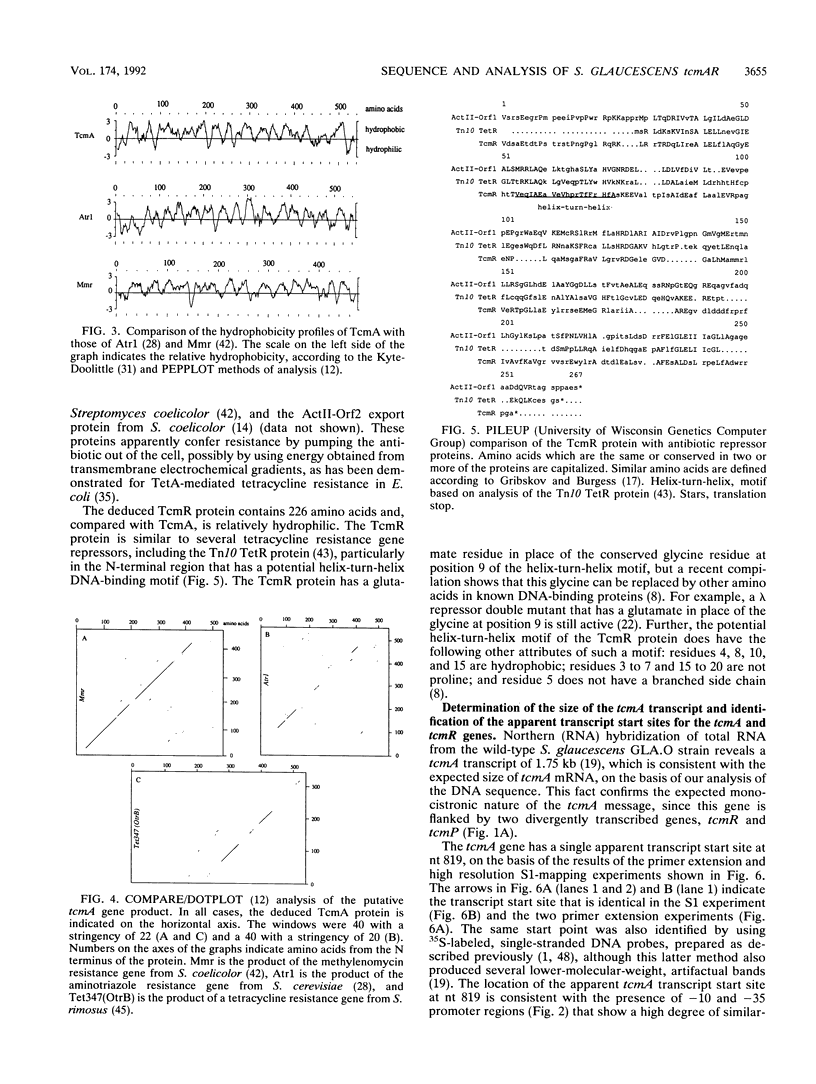
- Guilfoile P. G., Hutchinson C. R. The Streptomyces glaucescens TcmR protein represses transcription of the divergently oriented tcmR and tcmA genes by binding to an intergenic operator region. J Bacteriol. 1992 Jun;174(11):3659–3666. doi: 10.1128/jb.174.11.3659-3666.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochschild A., Irwin N., Ptashne M. Repressor structure and the mechanism of positive control. Cell. 1983 Feb;32(2):319–325. doi: 10.1016/0092-8674(83)90451-8. [DOI] [PubMed] [Google Scholar]
- Hopwood D. A. The Leeuwenhoek lecture, 1987. Towards an understanding of gene switching in Streptomyces, the basis of sporulation and antibiotic production. Proc R Soc Lond B Biol Sci. 1988 Nov 22;235(1279):121–138. doi: 10.1098/rspb.1988.0067. [DOI] [PubMed] [Google Scholar]
- Hoshiko S., Nojiri C., Matsunaga K., Katsumata K., Satoh E., Nagaoka K. Nucleotide sequence of the ribostamycin phosphotransferase gene and of its control region in Streptomyces ribosidificus. Gene. 1988 Sep 7;68(2):285–296. doi: 10.1016/0378-1119(88)90031-5. [DOI] [PubMed] [Google Scholar]
- Janssen G. R., Ward J. M., Bibb M. J. Unusual transcriptional and translational features of the aminoglycoside phosphotransferase gene (aph) from Streptomyces fradiae. Genes Dev. 1989 Mar;3(3):415–429. doi: 10.1101/gad.3.3.415. [DOI] [PubMed] [Google Scholar]
- Kanazawa S., Driscoll M., Struhl K. ATR1, a Saccharomyces cerevisiae gene encoding a transmembrane protein required for aminotriazole resistance. Mol Cell Biol. 1988 Feb;8(2):664–673. doi: 10.1128/mcb.8.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz E., Thompson C. J., Hopwood D. A. Cloning and expression of the tyrosinase gene from Streptomyces antibioticus in Streptomyces lividans. J Gen Microbiol. 1983 Sep;129(9):2703–2714. doi: 10.1099/00221287-129-9-2703. [DOI] [PubMed] [Google Scholar]
- Kumada Y., Anzai H., Takano E., Murakami T., Hara O., Itoh R., Imai S., Satoh A., Nagaoka K. The bialaphos resistance gene (bar) plays a role in both self-defense and bialaphos biosynthesis in Streptomyces hygroscopicus. J Antibiot (Tokyo) 1988 Dec;41(12):1838–1845. doi: 10.7164/antibiotics.41.1838. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Larson J. L., Hershberger C. L. The minimal replicon of a streptomycete plasmid produces an ultrahigh level of plasmid DNA. Plasmid. 1986 May;15(3):199–209. doi: 10.1016/0147-619x(86)90038-7. [DOI] [PubMed] [Google Scholar]
- Lee S. Y., Rasheed S. A simple procedure for maximum yield of high-quality plasmid DNA. Biotechniques. 1990 Dec;9(6):676–679. [PubMed] [Google Scholar]
- Li Y., Dosch D. C., Strohl W. R., Floss H. G. Nucleotide sequence and transcriptional analysis of the nosiheptide-resistance gene from Streptomyces actuosus. Gene. 1990 Jul 2;91(1):9–17. doi: 10.1016/0378-1119(90)90156-l. [DOI] [PubMed] [Google Scholar]
- McMurry L., Petrucci R. E., Jr, Levy S. B. Active efflux of tetracycline encoded by four genetically different tetracycline resistance determinants in Escherichia coli. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3974–3977. doi: 10.1073/pnas.77.7.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
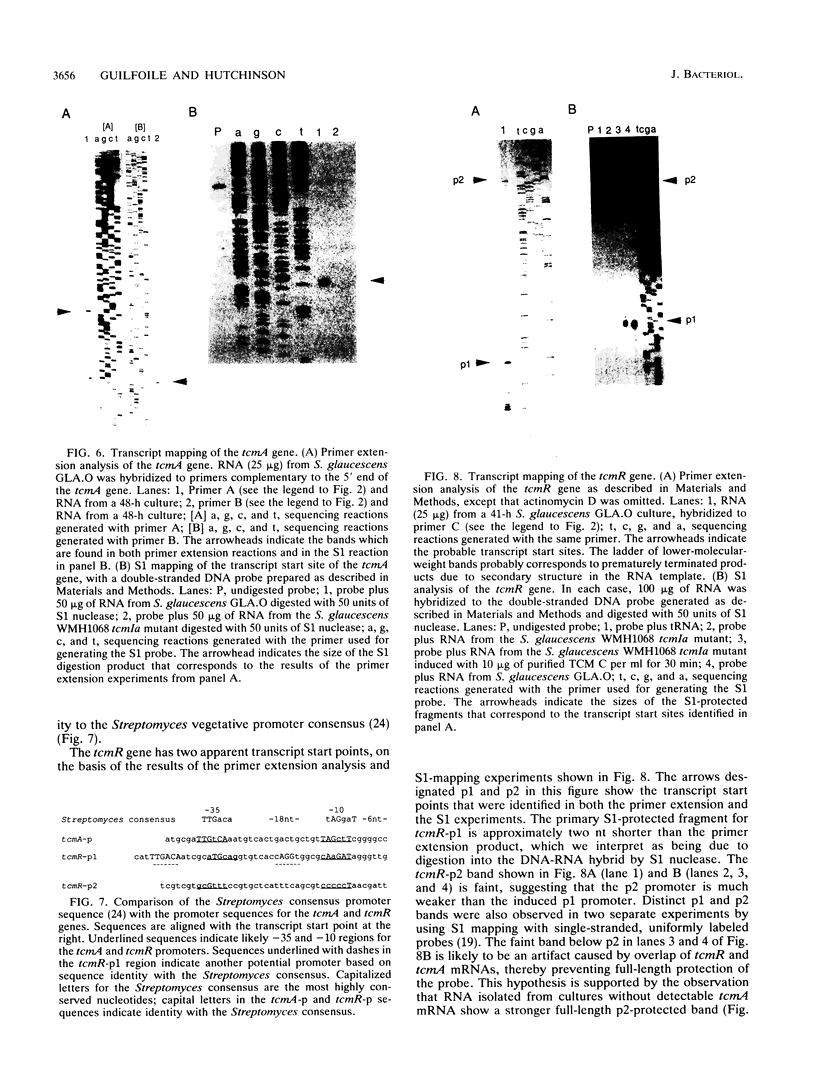
- Motamedi H., Hutchinson C. R. Cloning and heterologous expression of a gene cluster for the biosynthesis of tetracenomycin C, the anthracycline antitumor antibiotic of Streptomyces glaucescens. Proc Natl Acad Sci U S A. 1987 Jul;84(13):4445–4449. doi: 10.1073/pnas.84.13.4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motamedi H., Wendt-Pienkowski E., Hutchinson C. R. Isolation of tetracenomycin C-nonproducing Streptomyces glaucescens mutants. J Bacteriol. 1986 Aug;167(2):575–580. doi: 10.1128/jb.167.2.575-580.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray M. G. Use of sodium trichloroacetate and mung bean nuclease to increase sensitivity and precision during transcript mapping. Anal Biochem. 1986 Oct;158(1):165–170. doi: 10.1016/0003-2697(86)90605-6. [DOI] [PubMed] [Google Scholar]
- Neal R. J., Chater K. F. Nucleotide sequence analysis reveals similarities between proteins determining methylenomycin A resistance in Streptomyces and tetracycline resistance in eubacteria. Gene. 1987;58(2-3):229–241. doi: 10.1016/0378-1119(87)90378-7. [DOI] [PubMed] [Google Scholar]
- Postle K., Nguyen T. T., Bertrand K. P. Nucleotide sequence of the repressor gene of the TN10 tetracycline resistance determinant. Nucleic Acids Res. 1984 Jun 25;12(12):4849–4863. doi: 10.1093/nar/12.12.4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido D., Jiménez A. Optimization of gene expression in Streptomyces lividans by a transcription terminator. Nucleic Acids Res. 1987 May 26;15(10):4227–4240. doi: 10.1093/nar/15.10.4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynes J. P., Calmels T., Drocourt D., Tiraby G. Cloning, expression in Escherichia coli and nucleotide sequence of a tetracycline-resistance gene from Streptomyces rimosus. J Gen Microbiol. 1988 Mar;134(3):585–598. doi: 10.1099/00221287-134-3-585. [DOI] [PubMed] [Google Scholar]
- Rosteck P. R., Jr, Reynolds P. A., Hershberger C. L. Homology between proteins controlling Streptomyces fradiae tylosin resistance and ATP-binding transport. Gene. 1991 Jun 15;102(1):27–32. doi: 10.1016/0378-1119(91)90533-h. [DOI] [PubMed] [Google Scholar]
- Sharrocks A. D., Hornby D. P. S1 nuclease transcript mapping using sequenase-derived single-stranded probes. Biotechniques. 1991 Apr;10(4):426–428. [PubMed] [Google Scholar]
- Sheridan R. P., Chopra I. Origin of tetracycline efflux proteins: conclusions from nucleotide sequence analysis. Mol Microbiol. 1991 Apr;5(4):895–900. doi: 10.1111/j.1365-2958.1991.tb00763.x. [DOI] [PubMed] [Google Scholar]
- Stein D. S., Kendall K. J., Cohen S. N. Identification and analysis of transcriptional regulatory signals for the kil and kor loci of Streptomyces plasmid pIJ101. J Bacteriol. 1989 Nov;171(11):5768–5775. doi: 10.1128/jb.171.11.5768-5775.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauch E., Wohlleben W., Pühler A. Cloning of a phosphinothricin N-acetyltransferase gene from Streptomyces viridochromogenes Tü494 and its expression in Streptomyces lividans and Escherichia coli. Gene. 1988;63(1):65–74. doi: 10.1016/0378-1119(88)90546-x. [DOI] [PubMed] [Google Scholar]
- Thiara A. S., Cundliffe E. Interplay of novobiocin-resistant and -sensitive DNA gyrase activities in self-protection of the novobiocin producer, Streptomyces sphaeroides. Gene. 1989 Sep 1;81(1):65–72. doi: 10.1016/0378-1119(89)90337-5. [DOI] [PubMed] [Google Scholar]
- Turner D. H., Sugimoto N., Jaeger J. A., Longfellow C. E., Freier S. M., Kierzek R. Improved parameters for prediction of RNA structure. Cold Spring Harb Symp Quant Biol. 1987;52:123–133. doi: 10.1101/sqb.1987.052.01.017. [DOI] [PubMed] [Google Scholar]
- Uchiyama H., Weisblum B. N-Methyl transferase of Streptomyces erythraeus that confers resistance to the macrolide-lincosamide-streptogramin B antibiotics: amino acid sequence and its homology to cognate R-factor enzymes from pathogenic bacilli and cocci. Gene. 1985;38(1-3):103–110. doi: 10.1016/0378-1119(85)90208-2. [DOI] [PubMed] [Google Scholar]
- Vara J., Lewandowska-Skarbek M., Wang Y. G., Donadio S., Hutchinson C. R. Cloning of genes governing the deoxysugar portion of the erythromycin biosynthesis pathway in Saccharopolyspora erythraea (Streptomyces erythreus). J Bacteriol. 1989 Nov;171(11):5872–5881. doi: 10.1128/jb.171.11.5872-5881.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber W., Zähner H., Siebers J., Schröder K., Zeeck A. Stoffwechselprodukte von Mikroorganismen. 175. Mitteilung. Tetracenomycin C. Arch Microbiol. 1979 May;121(2):111–116. doi: 10.1007/BF00689973. [DOI] [PubMed] [Google Scholar]
- Westpheling J., Brawner M. Two transcribing activities are involved in expression of the Streptomyces galactose operon. J Bacteriol. 1989 Mar;171(3):1355–1361. doi: 10.1128/jb.171.3.1355-1361.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yue S., Motamedi H., Wendt-Pienkowski E., Hutchinson C. R. Anthracycline metabolites of tetracenomycin C-nonproducing Streptomyces glaucescens mutants. J Bacteriol. 1986 Aug;167(2):581–586. doi: 10.1128/jb.167.2.581-586.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]