Abstract
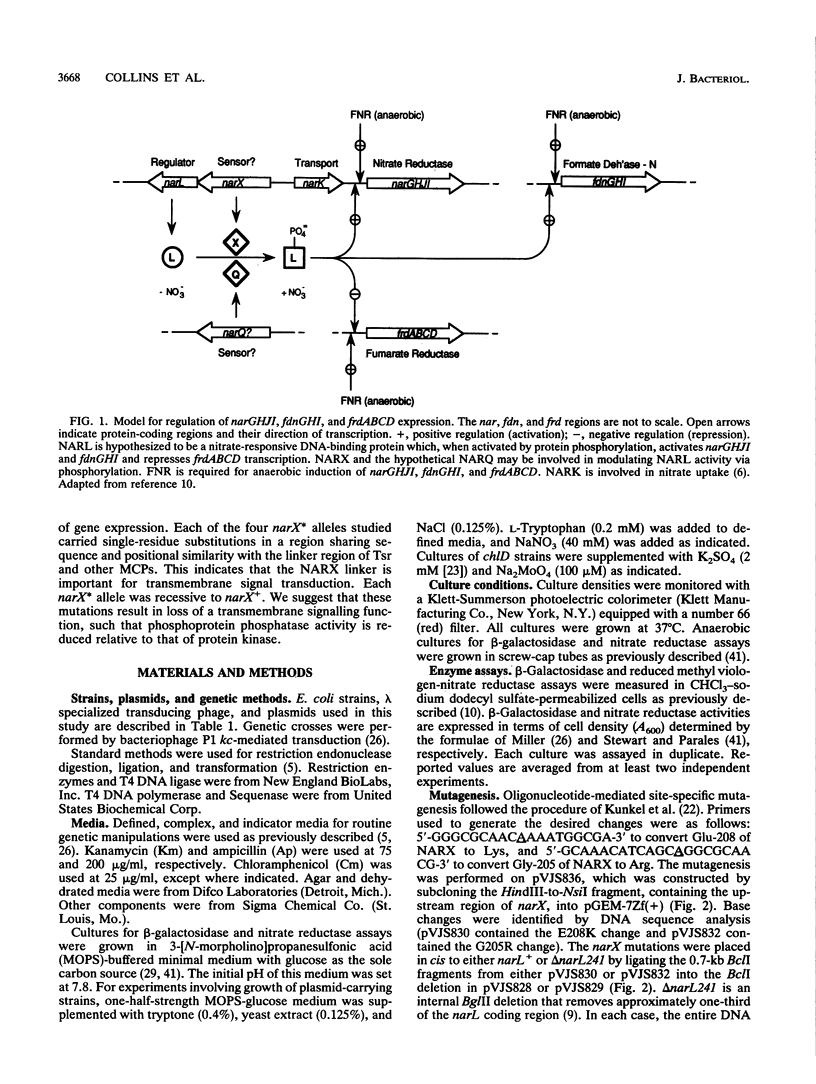
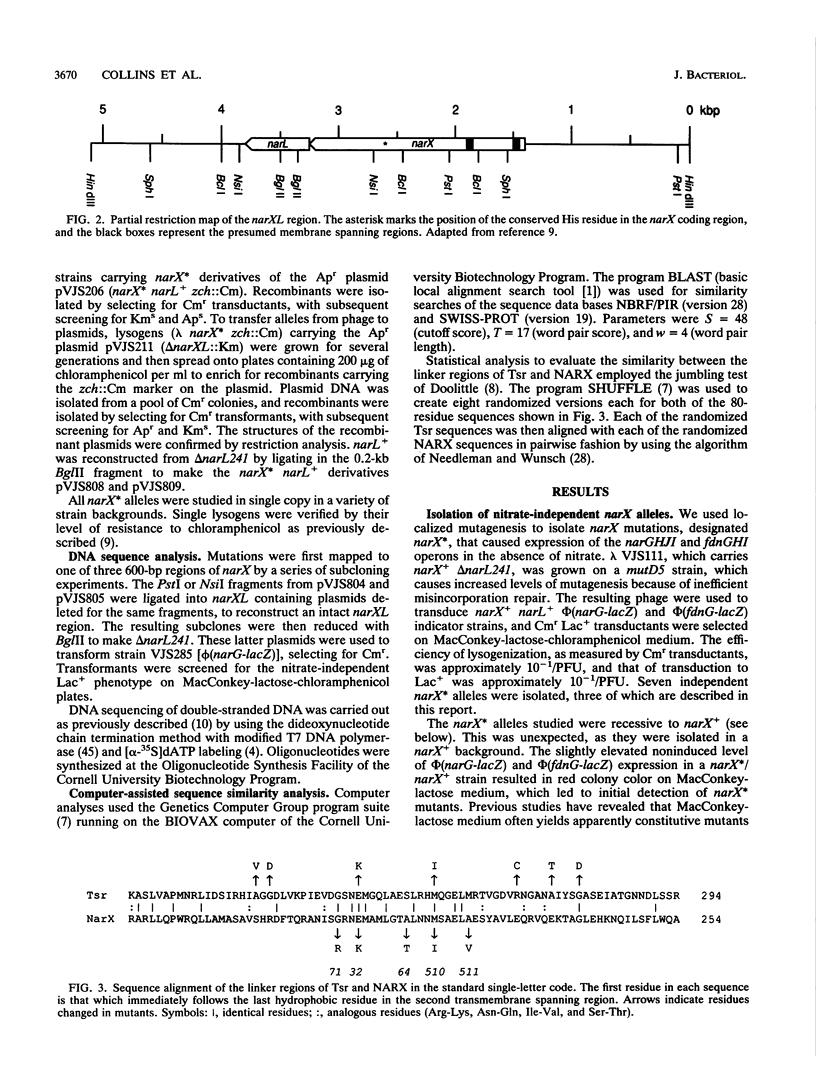
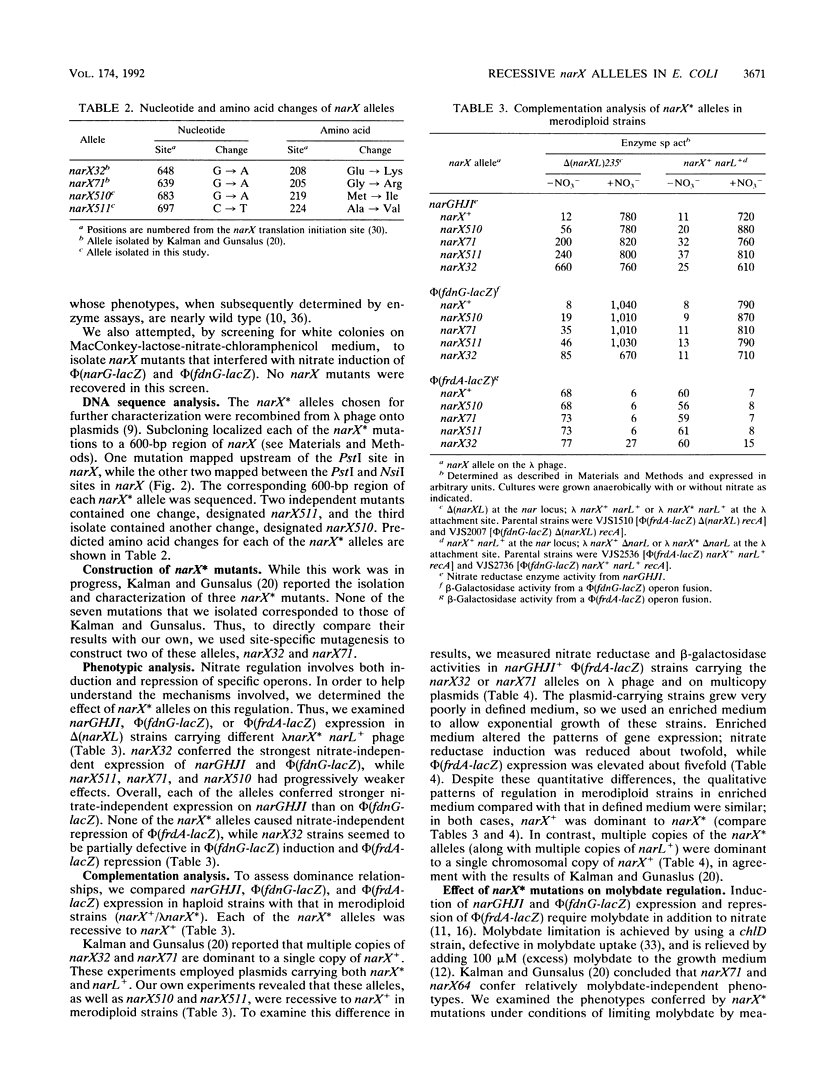
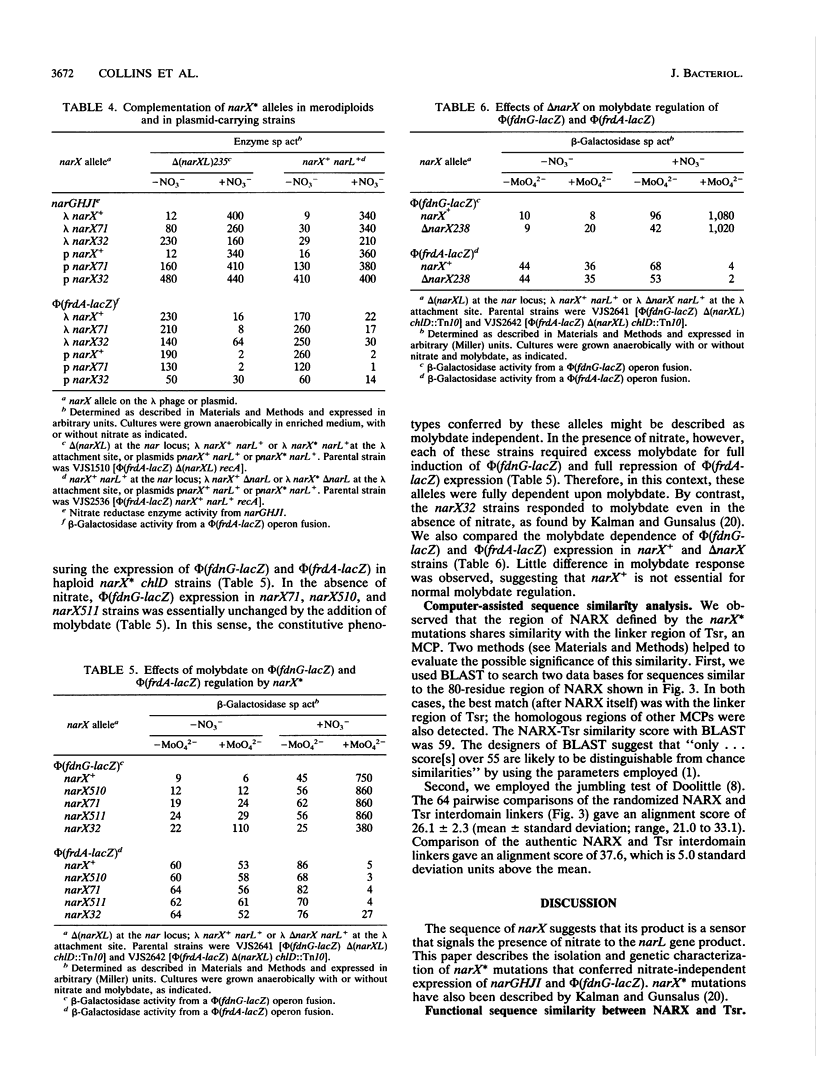
During anaerobic growth, nitrate induces synthesis of the anaerobic respiratory enzymes formate dehydrogenase-N and nitrate reductase. This induction is mediated by a transcription activator, the narL gene product. The narX gene product may be involved in sensing nitrate and phosphorylating NARL. We isolated narX mutants, designated narX*, that caused nitrate-independent expression of the formate dehydrogenase-N and nitrate reductase structural genes. We used lambda narX specialized transducing phage to genetically analyze these lesions in single copy. Two previously isolated narX* mutations, narX32 and narX71, were also constructed by site-specific mutagenesis. We found that each of these alleles caused nitrate-independent synthesis of formate dehydrogenase-N and nitrate reductase, and each was recessive to narX+. The narX* mutations lie in a region of similarity with the methyl-accepting chemotaxis protein Tsr. We suggest that the narX* proteins have lost a transmembrane signalling function such that phosphoprotein phosphatase activity is reduced relative to protein kinase activity.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ames P., Parkinson J. S. Transmembrane signaling by bacterial chemoreceptors: E. coli transducers with locked signal output. Cell. 1988 Dec 2;55(5):817–826. doi: 10.1016/0092-8674(88)90137-7. [DOI] [PubMed] [Google Scholar]
- Berg B. L., Stewart V. Structural genes for nitrate-inducible formate dehydrogenase in Escherichia coli K-12. Genetics. 1990 Aug;125(4):691–702. doi: 10.1093/genetics/125.4.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMoss J. A., Hsu P. Y. NarK enhances nitrate uptake and nitrite excretion in Escherichia coli. J Bacteriol. 1991 Jun;173(11):3303–3310. doi: 10.1128/jb.173.11.3303-3310.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan S. M., Stewart V. Mutational analysis of nitrate regulatory gene narL in Escherichia coli K-12. J Bacteriol. 1991 Jul;173(14):4424–4432. doi: 10.1128/jb.173.14.4424-4432.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan S. M., Stewart V. Nitrate regulation of anaerobic respiratory gene expression in narX deletion mutants of Escherichia coli K-12. J Bacteriol. 1990 Sep;172(9):5020–5029. doi: 10.1128/jb.172.9.5020-5029.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser J. H., DeMoss J. A. Phenotypic restoration by molybdate of nitrate reductase activity in chlD mutants of Escherichia coli. J Bacteriol. 1971 Nov;108(2):854–860. doi: 10.1128/jb.108.2.854-860.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunsalus R. P., Kalman L. V., Stewart R. R. Nucleotide sequence of the narL gene that is involved in global regulation of nitrate controlled respiratory genes of Escherichia coli. Nucleic Acids Res. 1989 Mar 11;17(5):1965–1975. doi: 10.1093/nar/17.5.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igo M. M., Ninfa A. J., Stock J. B., Silhavy T. J. Phosphorylation and dephosphorylation of a bacterial transcriptional activator by a transmembrane receptor. Genes Dev. 1989 Nov;3(11):1725–1734. doi: 10.1101/gad.3.11.1725. [DOI] [PubMed] [Google Scholar]
- Iuchi S., Lin E. C. Adaptation of Escherichia coli to respiratory conditions: regulation of gene expression. Cell. 1991 Jul 12;66(1):5–7. doi: 10.1016/0092-8674(91)90130-q. [DOI] [PubMed] [Google Scholar]
- Iuchi S., Lin E. C. Molybdenum effector of fumarate reductase repression and nitrate reductase induction in Escherichia coli. J Bacteriol. 1987 Aug;169(8):3720–3725. doi: 10.1128/jb.169.8.3720-3725.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S., Lin E. C. The narL gene product activates the nitrate reductase operon and represses the fumarate reductase and trimethylamine N-oxide reductase operons in Escherichia coli. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3901–3905. doi: 10.1073/pnas.84.11.3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalman L. V., Gunsalus R. P. Identification of a second gene involved in global regulation of fumarate reductase and other nitrate-controlled genes for anaerobic respiration in Escherichia coli. J Bacteriol. 1989 Jul;171(7):3810–3816. doi: 10.1128/jb.171.7.3810-3816.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalman L. V., Gunsalus R. P. Nitrate- and molybdenum-independent signal transduction mutations in narX that alter regulation of anaerobic respiratory genes in Escherichia coli. J Bacteriol. 1990 Dec;172(12):7049–7056. doi: 10.1128/jb.172.12.7049-7056.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalman L. V., Gunsalus R. P. The frdR gene of Escherichia coli globally regulates several operons involved in anaerobic growth in response to nitrate. J Bacteriol. 1988 Feb;170(2):623–629. doi: 10.1128/jb.170.2.623-629.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keener J., Kustu S. Protein kinase and phosphoprotein phosphatase activities of nitrogen regulatory proteins NTRB and NTRC of enteric bacteria: roles of the conserved amino-terminal domain of NTRC. Proc Natl Acad Sci U S A. 1988 Jul;85(14):4976–4980. doi: 10.1073/pnas.85.14.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lee J. H., Wendt J. C., Shanmugam K. T. Identification of a new gene, molR, essential for utilization of molybdate by Escherichia coli. J Bacteriol. 1990 Apr;172(4):2079–2087. doi: 10.1128/jb.172.4.2079-2087.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeil T., Roberts G. P., MacNeil D., Tyler B. The products of glnL and glnG are bifunctional regulatory proteins. Mol Gen Genet. 1982;188(2):325–333. doi: 10.1007/BF00332696. [DOI] [PubMed] [Google Scholar]
- Milligan D. L., Koshland D. E., Jr Intrasubunit signal transduction by the aspartate chemoreceptor. Science. 1991 Dec 13;254(5038):1651–1654. doi: 10.1126/science.1661030. [DOI] [PubMed] [Google Scholar]
- Needleman S. B., Wunsch C. D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970 Mar;48(3):443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Neidhardt F. C., Bloch P. L., Smith D. F. Culture medium for enterobacteria. J Bacteriol. 1974 Sep;119(3):736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohno T., Noji S., Taniguchi S., Saito T. The narX and narL genes encoding the nitrate-sensing regulators of Escherichia coli are homologous to a family of prokaryotic two-component regulatory genes. Nucleic Acids Res. 1989 Apr 25;17(8):2947–2957. doi: 10.1093/nar/17.8.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo F. D., Silhavy T. J. EnvZ controls the concentration of phosphorylated OmpR to mediate osmoregulation of the porin genes. J Mol Biol. 1991 Dec 5;222(3):567–580. doi: 10.1016/0022-2836(91)90497-t. [DOI] [PubMed] [Google Scholar]
- Scott D., Amy N. K. Molybdenum accumulation in chlD mutants of Escherichia coli. J Bacteriol. 1989 Mar;171(3):1284–1287. doi: 10.1128/jb.171.3.1284-1287.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slauch J. M., Silhavy T. J. Genetic analysis of the switch that controls porin gene expression in Escherichia coli K-12. J Mol Biol. 1989 Nov 20;210(2):281–292. doi: 10.1016/0022-2836(89)90330-6. [DOI] [PubMed] [Google Scholar]
- Slauch J. M., Silhavy T. J. cis-acting ompF mutations that result in OmpR-dependent constitutive expression. J Bacteriol. 1991 Jul;173(13):4039–4048. doi: 10.1128/jb.173.13.4039-4048.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart G. S., Lubinsky-Mink S., Jackson C. G., Cassel A., Kuhn J. pHG165: a pBR322 copy number derivative of pUC8 for cloning and expression. Plasmid. 1986 May;15(3):172–181. doi: 10.1016/0147-619x(86)90035-1. [DOI] [PubMed] [Google Scholar]
- Stewart V., Berg B. L. Influence of nar (nitrate reductase) genes on nitrate inhibition of formate-hydrogen lyase and fumarate reductase gene expression in Escherichia coli K-12. J Bacteriol. 1988 Oct;170(10):4437–4444. doi: 10.1128/jb.170.10.4437-4444.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart V. Nitrate respiration in relation to facultative metabolism in enterobacteria. Microbiol Rev. 1988 Jun;52(2):190–232. doi: 10.1128/mr.52.2.190-232.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart V., Parales J., Jr Identification and expression of genes narL and narX of the nar (nitrate reductase) locus in Escherichia coli K-12. J Bacteriol. 1988 Apr;170(4):1589–1597. doi: 10.1128/jb.170.4.1589-1597.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart V., Parales J., Jr, Merkel S. M. Structure of genes narL and narX of the nar (nitrate reductase) locus in Escherichia coli K-12. J Bacteriol. 1989 Apr;171(4):2229–2234. doi: 10.1128/jb.171.4.2229-2234.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart V. Requirement of Fnr and NarL functions for nitrate reductase expression in Escherichia coli K-12. J Bacteriol. 1982 Sep;151(3):1320–1325. doi: 10.1128/jb.151.3.1320-1325.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock J. B., Lukat G. S., Stock A. M. Bacterial chemotaxis and the molecular logic of intracellular signal transduction networks. Annu Rev Biophys Biophys Chem. 1991;20:109–136. doi: 10.1146/annurev.bb.20.060191.000545. [DOI] [PubMed] [Google Scholar]
- Stock J. B., Ninfa A. J., Stock A. M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989 Dec;53(4):450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss V., Magasanik B. Phosphorylation of nitrogen regulator I (NRI) of Escherichia coli. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8919–8923. doi: 10.1073/pnas.85.23.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. H., Clarke C. H., Marinus M. G. Specificity of Escherichia coli mutD and mutL mutator strains. Gene. 1990 Mar 1;87(1):1–5. doi: 10.1016/0378-1119(90)90488-d. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]


