Abstract
γ-Linolenic acid (GLA; C18:3 Δ6,9,12) is a component of the seed oils of evening primrose (Oenothera spp.), borage (Borago officinalis L.), and some other plants. It is widely used as a dietary supplement and for treatment of various medical conditions. GLA is synthesized by a Δ6-fatty acid desaturase using linoleic acid (C18:2 Δ9,12) as a substrate. To enable the production of GLA in conventional oilseeds, we have isolated a cDNA encoding the Δ6-fatty acid desaturase from developing seeds of borage and confirmed its function by expression in transgenic tobacco plants. Analysis of leaf lipids from a transformed plant demonstrated the accumulation of GLA and octadecatetraenoic acid (C18:4 Δ6,9,12,15) to levels of 13.2% and 9.6% of the total fatty acids, respectively. The borage Δ6-fatty acid desaturase differs from other desaturase enzymes, characterized from higher plants previously, by the presence of an N-terminal domain related to cytochrome b5.
Δ6-Desaturated fatty acids are of major importance in animal cells as they have roles in the maintenance of membrane structure and function, in the regulation of cholesterol synthesis and transport, in the prevention of water loss from the skin, and as precursors of eicosanoids, including prostaglandins and leucotrienes (1). In animals, members of this class of fatty acids are synthesized from the essential fatty acid linoleic acid (C18:2 Δ9,12), the first step being the desaturation to γ-linolenic acid (GLA; C18:3 Δ6,9,12) catalyzed by a Δ6-desaturase (1). Decreased activity of this key enzyme, observed for example in aging, stress, diabetes, eczema, and some infections, or increased catabolism of GLA resulting from oxidation or more rapid cell division (e.g., in cancer or inflammation) may lead to a deficiency of GLA (reviewed in ref. 2). Clinical trials have shown that dietary supplementation with GLA may be effective in treating a number of such conditions (e.g., atopic eczema, mastalgia, diabetic neuropathy, viral infections, and some types of cancer; ref. 2). Oils containing GLA are therefore widely used as a general health supplement and have been registered for pharmaceutical use.
In the plant kingdom, GLA is an uncommon fatty acid (3). Only a small number of higher plant species synthesize GLA, and in many of these, the fatty acid is found exclusively in the seed. GLA is also present in some fungi (e.g., Mucor javanicus) and cyanobacteria (3). Major commercial sources of GLA (4) are evening primrose (Oenothera spp.), in which GLA accounts for about 8–10% of the seed oil and borage (starflower) (Borago officinalis L.) seeds that contain some 20–25% GLA. These plants, however, suffer from poor agronomic performance and low yield; borage, for example, produces 300–600 kg/ha in the United Kingdom (4) compared with about 3 t/ha for oilseed rape. There is therefore considerable interest in both increasing the GLA content of existing crops and the production of GLA in a conventional oil crop (such as high linoleate rape).
In the higher plant cell, the synthesis of saturated fatty acids with chain lengths up to C18 and monounsaturated fatty acids (generally with a double bond at the Δ9 position) occurs in the plastid. Further desaturation can then occur either in the plastid or on the endoplasmic reticulum (ER; ref. 5). The desaturase enzymes of the plastid require reduced ferredoxin as an electron donor and are either soluble enzymes acting on saturated acyl-ACP substrates or membrane-bound enzymes using unsaturated fatty acids esterified to complex lipids such as monogalactosyldialglycerol. In contrast, the ER-located Δ12- and Δ15-desaturases use fatty acids located at the sn-2 position of phosphatidylcholine as substrates, and cytochrome b5 as a cofactor (5, 6). The Δ6-fatty acid desaturase in the developing cotyledons of borage is similar to the Δ12- and Δ15-desaturases in its location and substrate specificity (oleate/linoleate at the sn-2 position of phosphatidylcholine), and is assumed to use cytochrome b5 as its electron donor (7, 8). In addition, α-linolenic acid esterified to phosphatidylcholine may act as a substrate, resulting in the accumulation of octadecatetraenoic acid (OTA; C18:4 Δ6,9,12,15) in borage leaves (9).
We describe the isolation of a cDNA clone encoding the Δ6-fatty acid desaturase from developing seeds of borage, using a PCR-based strategy. The identity of the cDNA has been confirmed by functional expression and analysis in transgenic tobacco plants. The encoded protein differs from other membrane-bound fatty acid desaturases of plants, such as those encoded by the FAD genes of Arabidopsis (10, 11), in that the desaturase domain is preceded at the N terminus by a sequence that is related to cytochrome b5 (12), the haemprotein involved in electron transport to other ER-located fatty acid desaturases (Δ12,15) from higher plants (8, 13).
MATERIALS AND METHODS
Nucleic Acid Manipulations.
Total RNA was isolated from developing seeds of borage (B. officinalis) using guanidinium thiocyanate according to the method described in ref. 14. Poly(A)+ RNA was purified from total RNA using oligo(dT) cellulose according to standard methods (15) and was used as a template for cDNA library construction. Single-stranded cDNA was synthesized from total RNA using the Reverse Transcription System (Promega) according to the supplier’s instructions and used as a template for PCR amplification with degenerate primers. All nucleotide sequences were determined by the dideoxy chain termination method (15), and aligned using the gcg 8 program (16).
PCR-Based Cloning.
Two highly degenerate primers were synthesized for cDNA screening: forward primer A, 5′-GCGAATTC(A/G)TXGGXCA(T/C)GA(T/C)TG(T/C)GGXCA-3′ (fully degenerate to the conserved amino acid sequence GHDCGH), and reverse primer B, 5′-GCGAATTCATXT(G/T)XGG(A/G)AAXA(G/A)(A/G)TG(A/G)TG-3′ (fully degenerate to conserved amino acid sequence HHLFP), where X substitutes nucleotides AGTC. Each primer contained an EcoRI site (underlined) at the 5′ end to facilitate subsequent manipulations. These primers were used for PCR amplification with cDNA transcribed from total RNA. Reactions were run on a Perkin–Elmer Cetus DNA thermal cycler using a program of 1 min at 94°C, 1 min at 45°C, and 2 min at 72°C for 35 cycles followed by extension for 10 min at 72°C. PCR amplification products were separated on 1.0–2% agarose gels. PCR fragments of the expected length (600–700 bp) were purified using the Wizard DNA purification system (Promega), ligated into pGEM-T Vector according to the pGEM-T Vector Cloning Kit (Promega), and transformed into XL1-blue Escherichia coli cells. Plasmid DNA was purified and sequenced using the Promega miniprep system.
Library Screening.
Poly(A)+ mRNA from developing seeds of borage was used as the template for the synthesis of a cDNA library; custom synthesis and packaging being carried out by CLONTECH. The cDNA was inserted into the EcoRI site of the phage vector λ ZAPII, and the resultant DNA was packaged into phage particles. The cDNA bank contained 2.0 × 106 clones with an average insert size of 2.0 kb. Filter replicas of this library were hybridized with the labeled DNA probe pBdes1 and with a tobacco cDNA encoding cytochrome b5 (17). Radiolabeling of DNA and screening of phage libraries were conducted using standard techniques (15). The full-length cDNA clone pBdes6 was isolated and sequenced on both strands.
Northern Blot Analysis.
RNA was separated by electrophoresis through 1% formaldehyde agarose gel, transferred to nylon membrane (Hybond N, Amersham), and bound by exposure to UV light for 2 min. Probes were made from the cDNA clone pBdes6 by random priming (15). The filters were hybridized and washed as described in ref. 17 and then exposed to x-ray film at −80°C using an intensifying screen.
Plant Transformation.
To facilitate preparation of plant expression constructs, flanking SalI and SmaI restriction enzyme sites were added to the coding region of clone pBdes6 by PCR amplification. Two oligonucleotides were synthesized based on the pBdes6 coding sequence: primer C, 5′-GCGTCGACATGGCTGCTCAAATCAAG-3′ (annealing to the initiating methionine, indicated in boldface type), and primer D, 5′-GCCCGGGTTAACCATGAGTGTGAAG-3′ (annealing up to the complement of the stop codon, indicated in boldface type). The SalI (primer C) and SmaI (primer D) restriction sites are underlined. The PCR product was purified and subcloned into the vector pJD330 (18) to generate the plasmid p35Bdes6. Digestion of p35Bdes6 with XbaI released fragment of ≈2,200 bp containing the ORF of the borage pBdes6, together with regulatory elements consisting of the cauliflower mosaic virus 35S promoter, an Ω-translational enhancer from tobacco mosaic virus (19) and the nopaline synthase (nos) termination sequence. This XbaI fragment was gel purified and cloned into pBIN19 (20) to obtain the plasmid pNTdes6, which was transformed into Agrobacterium tumefaciens strain LBA4404 by electroporation. Tobacco (Nicotiana tabacum cv. NVS) was transformed with the plant expression plasmid according to standard procedures (21). Initial transformants were selected on 50 μg/ml kanamycin and then transferred to 100 μg/ml kanamycin. Plants were maintained in axionic culture under controlled conditions.
Fatty Acid Analysis.
Lipids were extracted from leaves of transformed and control tobacco plants by homogenization in MeOH-CHCl3 using a modification of the method of Bligh and Dyer (22). The resulting CHCl3 phase was evaporated to dryness under nitrogen gas, and the samples were transmethylated with 1 M HCl in methanol at 80°C for 1 h. Fatty acid methyl esters (FAMes) were extracted in hexane and purified using a small column packed with Florisil. Analysis of FAMes was conducted using a Hewlett Packard 5880A Series Gas Chromatograph equipped with a 25 M × 0.32 mm RSL-500BP bonded capillary column and a flame ionization detector. Fatty acids were identified by comparison of retention times with FAMe standards (Sigma) separated on the same GC. Quantitation was carried out using peak height area integrals expressed as a total of all integrals.
GC–Mass Spectrometry (MS) Analysis.
Fatty acid 4,4-Dimethyloxazoline (DMOX) derivatives were prepared for GC-MS analysis by a modification of the method of Fay and Richli (23). Lipid samples (extracted from tobacco leaves as described above) were heated at 180°C in 2-amino-2-methyl-1-propanol under N2 for 18 h. After cooling to room temperature dichloromethane and water were added. The DMOX derivatives were recovered in the dichloromethane, passed through a column of anhydrous sodium sulfate to remove water, and dried under a stream of N2. To remove any contaminating polar material, the samples were taken up in hexane, passed through a short Florisil column, and evaporated to dryness. The samples were then dissolved in an appropriate volume of hexane for GC-MS analysis. Fatty acid DMOX derivatives were analyzed by GC-MS on a Hewlett Packard 5890 Series II Plus gas chromatograph equipped with a 50 M × 0.25 mm BPX70TM capillary column connected directly to a Hewlett Packard 5989B MS Engine quadropole mass spectrometer operating at an ionization energy of 70 eV and emission current of 300 μA. Mass spectra were interpreted by comparison to the mass spectra of DMOX derivatives of GLA and OTA prepared from blackcurrant oil, which is known to contain both these fatty acids (24), using the interpretation rules of ref. 25.
RESULTS
PCR-Based Cloning of Membrane-Bound Desaturases.
Comparisons of the deduced amino acid sequences of membrane-bound fatty acid desaturases (and related proteins) from mammals, fungi, insects, higher plants, and cyanobacteria reveal three highly conserved regions (boxes) containing histidine residues (26). Since the borage seed Δ6-desaturase is membrane-bound (27), two highly degenerate primers were constructed based on the sequences of the first and third histidine boxes present in the membrane-bound Δ12- and Δ15-fatty acid desaturases of plants. These primers were used in PCRs with cDNA transcribed from total RNA of developing cotyledons of borage. PCR products of the predicted length (600–700 bp) were cloned and sequenced, allowing them to be classified into three groups: 45% showed similarity to other proteins (i.e., not fatty acid desaturases), 35% resembled Δ12-desaturases, and 20% formed a separate group that showed some similarity to both Δ12- and Δ15-desaturases but was clearly distinct from the second group.
Sequencing of a representative clone (pBdes1) from the third group revealed an ORF of 228 aa with three putative histidine boxes. Alignment of the deduced amino acid sequence with those of known desaturases (data not shown) showed highest similarity to the Δ15-desaturases, although the actual level of identity was low (less than 30%). Since borage seed oil contains little or no α-linolenic acid, it is unlikely that high levels of transcripts for Δ15-desaturases would be present in the developing seeds. It was therefore considered likely that the pBdes1 PCR product encoded part of a putative Δ6-desaturase.
To isolate a full-length clone corresponding to the pBdes1 PCR product, the insert was used to probe a borage developing seed cDNA library constructed in λ ZAPII. A total of 3 × 105 plaques were screened, and 20 individual phage clones that hybridized with the pBdes1 DNA probe were identified and purified by further rounds of hybridization. Restriction enzyme digestion of 15 clones recovered from positive plaques showed the presence of single inserts that hybridized with the probe, ranging from 700 to 1,800 bp in length. One of these, termed pBdes6, containing an insert of 1,800 bp, was chosen for detailed analysis.
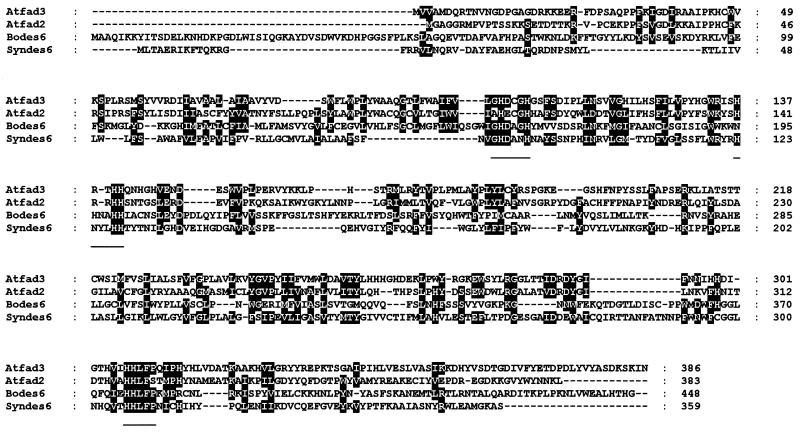
pBdes6 encodes a 1,344-bp ORF, preceded by a 41-bp 5′ untranslated region. The coding region was followed by a stop codon and a 345-bp untranslated region with a poly(A) tail. The ORF encoded 448 aa, corresponding to a putative protein with an Mr of about 50,000, which is significantly larger than the predicted Mr of other microsomal desaturases such as the Δ12- and Δ15-desaturases from Arabidopsis (refs. 10 and 11; Fig. 1). A degree of similarity to other fatty acid desaturases is clear, but only over a part of the coding sequence. The amino acid sequence from residues 144 to 448 showed about 17% identity with Δ15 (FAD3) (10) and Δ12 (FAD2) (10) desaturases from Arabidopsis and about 22% identity with a Δ6-desaturase from the cyanobacterium Synechocystis (28). The whole sequence was also 60% identical to a cDNA clone of unknown function isolated from sunflower seeds (29). The three conserved histidine boxes that are characteristic of other membrane-bound desaturases were also present, and located at similar positions within the sequence. The distance between the first and second boxes was 32 aa, compared with 31 or 32 aa in Δ12- and Δ15-desaturases, and the distance between the second and third boxes was 172 aa, compared with 132–173 in other membrane-bound desaturases. The importance of these histidine boxes in catalysis has been demonstrated by site-directed mutagenesis of the soluble Δ9-desaturase from rat and Δ12-desaturase of Synechocystis (26, 30).
Figure 1.
Comparison of the deduced amino acid sequence of pBdes6 (labeled bodes6) with other desaturases. The entire coding sequence of pBdes6 was compared with the Arabidopsis FAD2 (atfad2) and FAD3 (atfad3) microsomal desaturases, as well as the cyanobacterial Δ6 desaturase (syndes6). Identical or conserved residues are boxed, and the conserved histidine boxes are underlined. The sequence of pBdes6 has been deposited in the GenBank database (accession no. U79010U79010).
pBdes6 Encodes a Protein Containing a Cytochrome b5-Like Heme-Binding Domain.
The predicted hydrophobicity plot for the protein encoded by pBdes6 revealed a profile characteristic of a fatty acid desaturase, with the histidine boxes located in hydrophilic areas and separated by hydrophobic domains (not shown). The borage protein, however, contained a hydrophilic region at the N terminus longer than those of the membrane-bound Δ12- and Δ15-desaturases. Closer analysis showed significant sequence similarity between the first 90–100 aa at the N terminus of the protein encoded by pBdes6 and microsomal cytochrome b5 proteins from higher plants (17). This similarity included the presence of seven of the eight invariant residues of the cytochrome b5 class of proteins identified by Lederer (12). A heme-containing electron donor is required for fatty acid desaturation, and cytochrome b5 is known to fulfill this function with membrane-bound fatty desaturases (Δ12 and Δ15; refs. 8 and 13) and with the related Δ12-hydroxylase (31). We therefore isolated a cDNA for cytochrome b5 from the borage cDNA library using a tobacco cDNA (17) as a probe. Sequencing of this cDNA revealed an ORF encoding 132 aa which had some 80% sequence identity to cytochrome b5 proteins from tobacco and rice (17). It also showed 32% sequence identity with the cytochrome b5-related domain of the protein encoded by pBdes6 (Fig. 2). The identity is particularly high in regions previously identified as essential for cytochrome b5 function, including the EHPGG motif in the heme-binding region.
Figure 2.
Comparison of the deduced amino acid sequence of pBdes6 with plant cytochrome b5 sequences. The first 196 residues of pBdes6 (bodes6) were compared with cytochrome b5 sequences from borage (bocytb5), rice (oscytb5), and tobacco (ntcytb5). The conserved heme-binding domain is underlined. The sequence of borage cytochrome b5 has been deposited in the GenBank database (accession no. U79011U79011).
Functional Analysis of pBdes6 in Transgenic Tobacco.
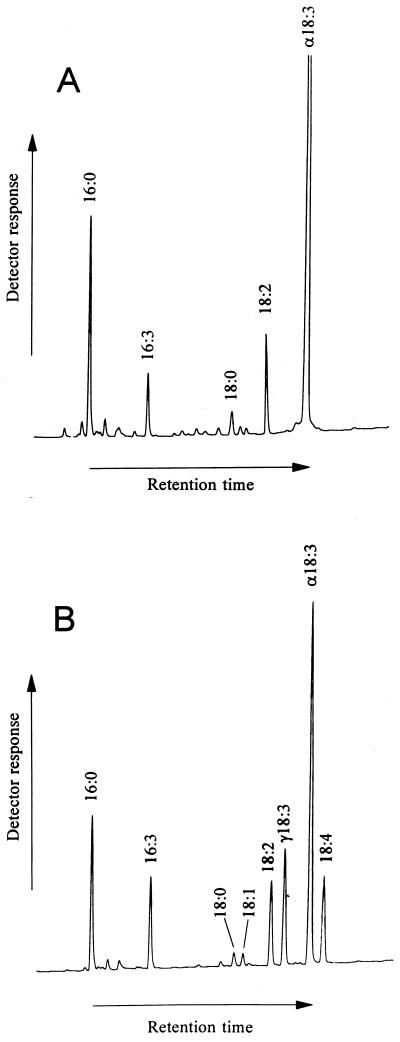
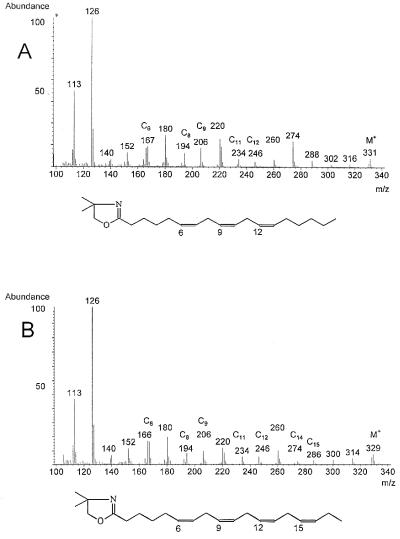
To confirm the identity of pBdes6 as a Δ6-fatty acid desaturase, the cDNA was transferred to tobacco plants under the control of an Ω-enhanced cauliflower mosaic virus 35S promoter via Agrobacterium-mediated gene transfer. Single leaves were removed from transformed and control plants, and FAMes were prepared from total lipid extracts and analyzed by GC (Fig. 3). Two peaks were observed in the chromatogram of FAMes from the transformants (Fig. 3B) not present in the control plants (Fig. 3A). These peaks had retention times identical to the FAMe standards of GLA (C18:3 Δ6,9,12) and OTA (C18:4 Δ6,9,12,15). Further analysis of the transformant by GC-MS analysis of fatty acid DMOX derivatives confirmed the identities of these peaks (Fig. 4). Both spectra contained abundant m/z 113 (McLafferty rearrangement ion) and 126 peaks typical of fatty acid DMOX derivatives (23). The spectrum of the putative GLA derivative (Fig. 4A) had a molecular ion at m/z 331, suggesting an octadecatrienoic fatty acid; gaps of 12 amu between m/z 194 and 206, and m/z 234 and 246, indicating double bonds at C9 and C12; and a prominent m/z 166/167 pair specific for a C6 double bond (25). The spectrum of the putative OTA (Fig. 4B) had an additional gap of 12 amu between m/z 274 and 286, indicating the presence of a C15 double bond.
Figure 3.
Identification of GLA and OTA in transgenic tobacco by GC. Chromatograms of FAMes from leaf tissue of control tobacco plant (A) or plant transformed with pBdes6 (B). Two novel peaks are seen in B; these peaks have retention times identical to FAMe standards of GLA and OTA. The identity of peaks (as determined by comparison of retention times with those of known standards) is indicated. Detection was by flame ionization.
Figure 4.
Mass spectra of DMOX-derivatized fatty acids. Spectra of the fatty acids identified in Fig. 3 as GLA (A) and OTA (B). Details of the interpretation of the spectra are given in the text. The deduced structures of the fatty acid derivatives are shown.
The proportions of fatty acids in the total lipid fractions prepared from the leaves of the control and transformed tobacco plants are given in Table 1. GLA and OTA account for about 13% and 10% of the total, respectively, in the transgenic material and are absent in the control plants. The presence of both GLA and OTA indicates that the Δ6-desaturase used both linoleic acid and α-linolenic acid as substrates, and this may be responsible for the decrease in α-linolenic acid observed in the transgenic line.
Table 1.
Total fatty acid content of lipid extracts from leaves of a control tobacco plant and a plant transformed with the borage Δ6 desaturase clone pBdes6
| Acids | % Fatty acid
|
|
|---|---|---|
| Control | Transformant | |
| Palmitic(C16:0) | 16.3 | 14.0 |
| Palmitoleic(C16:1) | Trace | Trace |
| (C16:3) | 5.0 | 9.0 |
| Stearic(C18:0) | 2.4 | 1.5 |
| Oleic(C18:1) | Trace | 1.3 |
| Linoleic(C18:2) | 9.1 | 9.5 |
| γ-Linolenic(C18:3) | ND | 13.2 |
| α-Linolenic (C18:3) | 65.1 | 40.1 |
| OTA(C18:4) | ND | 9.6 |
Percentages were integrated from peak areas of GC traces shown in Fig. 3. ND, not detected.
Northern Blot Analysis.
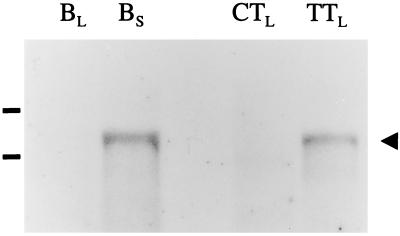
To provide further evidence that the introduction of the borage cDNA into the tobacco genome was responsible for these novel desaturation products, total RNA was isolated from the leaves of either a GLA-positive transgenic tobacco plant or a control plant that had been subject to the same tissue culture regime. RNA was also isolated from developing borage seeds and leaves, and the samples were analyzed by Northern blotting and probed with the pBdes6 cDNA (Fig. 5). A positive hybridization signal of identical mobility was obtained from RNA isolated from borage seeds and the transgenic GLA-positive tobacco line, but not from the control tobacco plant. Prolonged exposure of the autoradiograph showed that low levels of the pBdes6 transcript (or related transcripts) were present in the RNA samples extracted from borage leaves, a result that is consistent with the observed accumulation of GLA in the leaves of this species (9).
Figure 5.
Northern blot analysis of pBdes6 expression in borage and in transgenic tobacco. Total RNA (10 μg), extracted from borage leaves (BL), borage seeds (BS), control tobacco leaves (CTL) or transgenic tobacco leaves (TTL) was probed with 32P-labeled pBdes6. After hybridization and high stringency washing, the resulting autoradiograph indicated expression of the pBdes6 transcript (≈2,000 bp; marked with the arrowhead) in borage seeds and transgenic tobacco leaves. The positions of the rRNA bands are indicated.
DISCUSSION
We undertook to isolate a cDNA encoding a Δ6-desaturase from borage using a degenerate PCR approach based on conserved amino acid sequence motifs in other microsomal fatty acid desaturases (26, 30). Previous studies (9, 27) had shown that the borage Δ6-desaturase activity was associated with the microsomal membrane fraction and probably used cytochrome b5 as an electron donor, like the microsomal Δ12 (FAD2) and Δ15 (FAD3) desaturases. The borage cDNA clone (pBdes6) was confirmed to encode a Δ6-desaturase by ectopic expression in the leaves of transgenic tobacco, resulting in the accumulation of the fatty acids GLA and OTA. The borage Δ6-desaturase encoded by pBdes6 differed from previously characterized fatty acid desaturases from higher plants by the presence of an N-terminal extension related to the cytochrome b5 class of heme-binding proteins (13). This domain is not present in the plant microsomal Δ12- and Δ15-desaturases (10, 11) or in the related Δ12-hydroxylase (31) which have been cloned and functionally characterized in transgenic plants, although their use of microsomal cytochrome b5 as an electron donor has been clearly demonstrated (8, 32). It is also clear that the N-terminal cytochrome b5-related domain of pBdes6 is structurally distinct from the borage microsomal cytochrome b5, as it does not contain the conserved hydrophobic C-terminal microsomal membrane anchor normally present in cytochrome b5 proteins (17, 33). Since cytochrome b5 usually functions in association with the ER membrane, it is likely that the fusion protein described in this study has the same location and this is supported by the absence of any domains resembling chloroplast targeting transit sequences (34). Although the protein encoded by pBdes6 does not appear to have an N-terminal cleavable ER-targeting signal sequence (as judged by computer searching), the hydrophobic regions present in the protein would be sufficient to allow it associate with the endomembrane system. No obvious ER-retention motifs are present, but a potential glycosylation site is present at residues 278–280 (N-V-S).
Domains related to cytochrome b5 are also present in a microsomal Δ9-fatty acid desaturase (Ole1p, the OLE1 gene product) from yeast (35) and in other oxido/reductase enzymes (e.g., nitrate reductase, sulfite oxidase, and flavocytochrome b2; ref. 12). In the yeast Δ9-desaturase, this cytochrome b5 domain exists as a 113-aa C-terminal fusion. Expression of OLE1 from a multicopy plasmid rescued yeast double mutants that lacked both OLE1 and microsomal cytochrome b5 genes, unlike rescue by a rat microsomal Δ9-desaturase, which required the presence of the cytochrome b5 gene (35). Moreover, when the C-terminal b5 domain of OLE1 was deleted, the yeast cells remained fatty acid auxotrophic, even in the presence of endogenous yeast cytochrome b5, indicating that cytochrome b5 is not able to act in trans to complement the loss of the cytochrome b5 fusion domain of Ole1p (35). This suggests that the fusion domain plays an essential role in the desaturase reaction of this enzyme. A cDNA clone encoding a related cytochrome b5 fusion protein has also been isolated from sunflower seeds (29), as noted above, but the corresponding protein has not been identified. In the sunflower protein, the cytochrome b5 domain is fused to the N terminus of a putative desaturase sequence, as in the pBdes6 protein, and expression of this domain (≈120 residues) in E. coli (29) has shown that it is capable of undergoing reversible oxidation and reduction, indicating a functional heme group. Similar results were also obtained by expression of a tobacco cytochrome b5 cDNA in E. coli (33). However, sunflower seeds do not accumulate GLA (3) and would therefore not be expected to possess an active Δ6-desaturase. The substrate specificity of this sunflower protein is not known, and its role in fatty acid desaturation/hydroxylation-type reactions can only be inferred from sequence homology. Similarly, the functional and evolutionary significance of the existence of two types of membrane-bound desaturases in plants is not clear, although it can be suggested that the fusion of a cytochrome b5 domain to the desaturase may facilitate a more efficient electron transfer. It is also unclear why the yeast Ole1p desaturase has a C-terminal cytochrome b5 domain, whereas the borage desaturase has an N-terminal cytochrome b5 domain.
Recently, GLA and OTA accumulation in transgenic plants has been reported by Reddy and Thomas (36), who expressed a cyanobacterial Δ6-desaturase gene in tobacco. The combined levels of GLA and OTA varied from about 2% to 4% of the leaf C:18 fatty acids, with only small differences depending on whether the protein was targeted to the plastid, cytoplasm, or ER lumen. This low level of activity is perhaps not surprising as the cyanobacterial Δ6-desaturase differs from the ER-located higher plant desaturases in using ferredoxin rather than cytochrome b5 as a cofactor. The cyanobacterial Δ6-desaturase also resulted in the accumulation of comparatively higher levels of OTA than GLA, but the reason for this is not known. The levels of GLA and OTA accumulating in the leaves of transgenic tobacco plants expressing the borage desaturase encoded by pBdes6 account together for over 23% of total fatty acids, indicating the potential for producing GLA in transgenic oil crops. Sunflower would be particularly suitable in this respect as the presence of between 50% and 70% linoleic acid and with little or no α-linolenic acid (37) should facilitate the synthesis of high levels of GLA.
Acknowledgments
IACR receives grant-aided support from the Biotechnology and Biological Sciences Research Council of the United Kingdom. The work was supported by a grant from the Biotechnology and Biological Sciences Research Council under the Collaboration with Industry Scheme.
ABBREVIATIONS
- GLA
γ-linolenic acid
- OTA
octadecatetraenoic acid
- ER
endoplasmic reticulum
- FAMe
fatty acid methyl ester
- DMOX
4,4-dimethyloxazoline
- MS
mass spectrometry
Footnotes
References
- 1.Horrobin D F. Prog Lipid Res. 1992;31:163–194. doi: 10.1016/0163-7827(92)90008-7. [DOI] [PubMed] [Google Scholar]
- 2.Horrobin D F. Rev Contemp Pharmacother. 1990;1:1–45. [Google Scholar]
- 3.Gunstone F D. Prog Lipid Res. 1992;31:145–161. doi: 10.1016/0163-7827(92)90007-6. [DOI] [PubMed] [Google Scholar]
- 4.Fieldsend A F. Biologist. 1995;42:203–207. [Google Scholar]
- 5.Heinz E. In: Lipid Metabolism in Plants. Moore T S, editor. Boca Raton, FL: CRC; 1993. pp. 34–89. [Google Scholar]
- 6.Stymne S, Stobart A K. In: Seed Storage Compounds. Shewry P R, Stobart A K, editors. Oxford: Oxford Univ. Press; 1993. pp. 96–114. [Google Scholar]
- 7.Griffiths G, Brechany E Y, Christie W W, Stymne S, Stobart A K. In: Biological Role of Plant Lipids. Biacs P A, Gruiz K, Kremmer T, editors. New York: Plenum; 1989. pp. 151–154. [Google Scholar]
- 8.Smith M A, Cross A R, Jones O T G, Griffiths W T, Stymne S, Stobart A K. Biochem J. 1990;272:23–29. doi: 10.1042/bj2720023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Griffiths G, Brechany E Y, Jackson F M, Christie W W, Stymne S, Stobart A K. Phytochemistry. 1996;43:381–386. [Google Scholar]
- 10.Arondel V, Lemieux B, Hwang I, Gibson S, Goodman H, Somerville C R. Science. 1992;258:1353–1355. doi: 10.1126/science.1455229. [DOI] [PubMed] [Google Scholar]
- 11.Okuley J, Lightner J, Feldmann K, Yadav N, Lark E, Browse J. Plant Cell. 1994;6:147–158. doi: 10.1105/tpc.6.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lederer F. Biochimie. 1994;76:674–692. doi: 10.1016/0300-9084(94)90144-9. [DOI] [PubMed] [Google Scholar]
- 13.Kerns E V, Hugly S, Somerville C R. Arch Biochem Biophys. 1991;284:431–436. doi: 10.1016/0003-9861(91)90319-e. [DOI] [PubMed] [Google Scholar]
- 14.Napier J A, Smith M A, Stobart A K, Shewry P R. Planta. 1995;197:200–202. doi: 10.1007/BF00239957. [DOI] [PubMed] [Google Scholar]
- 15.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 16.Devereux J, Haeberli P, Smithies O. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith M A, Stobart A K, Shewry P R, Napier J A. Plant Mol Biol. 1994;25:527–537. doi: 10.1007/BF00043880. [DOI] [PubMed] [Google Scholar]
- 18.Zakai N, Ballas N, Hershkovitz M, Broido S, Ram R, Loyter A. Plant Mol Biol. 1993;21:823–830. doi: 10.1007/BF00027114. [DOI] [PubMed] [Google Scholar]
- 19.Gallie D R, Lucas W J, Walbot V. Plant Cell. 1989;1:301–311. doi: 10.1105/tpc.1.3.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bevan M. Nucleic Acid Res. 1984;12:8711–8722. doi: 10.1093/nar/12.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Draper J, Scott R, Armatige P, Walden R. Plant Genetic Transformation and Gene Expression: A Laboratory Manual. Oxford: Blackwell Scientific; 1988. [Google Scholar]
- 22.Bligh E G, Dyer W J. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 23.Fay L, Richli U. J Chromatogr. 1991;541:89–98. [Google Scholar]
- 24.Traitler H, Willie H J, Studer A. J Am Oil Chem Soc. 1988;65:755–760. [Google Scholar]
- 25.Zhang J Y, Yu Q T, Liu B N, Huang Z H. Biomed Environ Mass Spectrom. 1988;15:33–44. [Google Scholar]
- 26.Shanklin J, Whittle E, Fox B G. Biochemistry. 1994;33:12787–12794. doi: 10.1021/bi00209a009. [DOI] [PubMed] [Google Scholar]
- 27.Stymne S, Stobart A K. Biochem J. 1986;24:385–393. doi: 10.1042/bj2400385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reddy A S, Nuccio M L, Gross L M, Thomas T L. Plant Mol Biol. 1993;22:293–300. doi: 10.1007/BF00014936. [DOI] [PubMed] [Google Scholar]
- 29.Sperling P, Schmidt H, Heinz E. Eur J Biochem. 1995;232:798–805. [PubMed] [Google Scholar]
- 30.Avelange-Macherel M-H, Macherel D, Wada H, Murata N. FEBS Lett. 1995;361:111–114. doi: 10.1016/0014-5793(95)00163-4. [DOI] [PubMed] [Google Scholar]
- 31.van de Loo F J, Broun P, Turner S, Somerville C. Proc Natl Acad Sci USA. 1995;92:6743–6747. doi: 10.1073/pnas.92.15.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith M A, Jonsson L, Stymne S, Stobart A K. Biochem J. 1992;287:141–144. doi: 10.1042/bj2870141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith M A, Napier J A, Stymne S, Tatham A S, Shewry P R, Stobart A K. Biochem J. 1994;303:73–79. doi: 10.1042/bj3030073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Napier J A. In: Methods in Molecular Biology. Jones H, editor. Vol. 49. Totowa, NJ: Humana; 1995. pp. 369–376. [DOI] [PubMed] [Google Scholar]
- 35.Mitchell A G, Martin C E. J Biol Chem. 1995;270:29766–29772. doi: 10.1074/jbc.270.50.29766. [DOI] [PubMed] [Google Scholar]
- 36.Reddy A V, Thomas T L. Nat Biotechnol. 1996;14:639–642. doi: 10.1038/nbt0596-639. [DOI] [PubMed] [Google Scholar]
- 37.Salunkhe D K, Chavan J K, Adsule R N, Kadam S S. World Oilseeds—Chemistry, Technology and Utilization. New York: Van Nostrand Reinhold; 1991. [Google Scholar]