Abstract
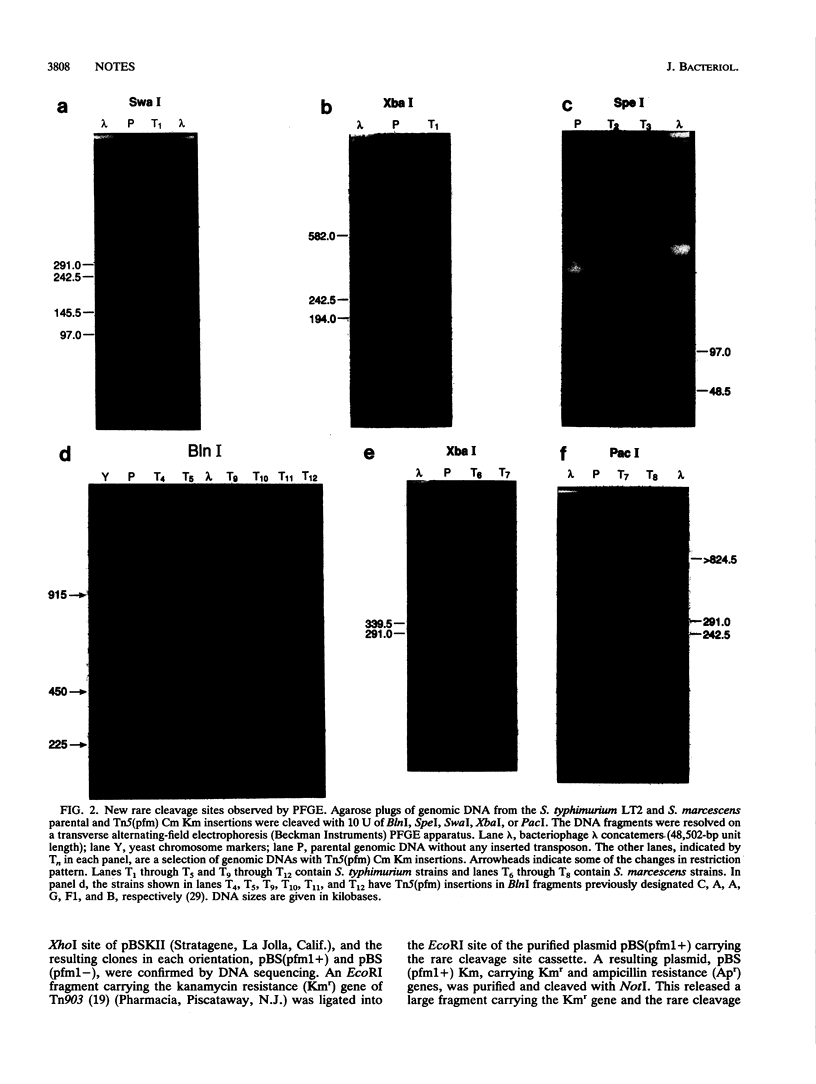
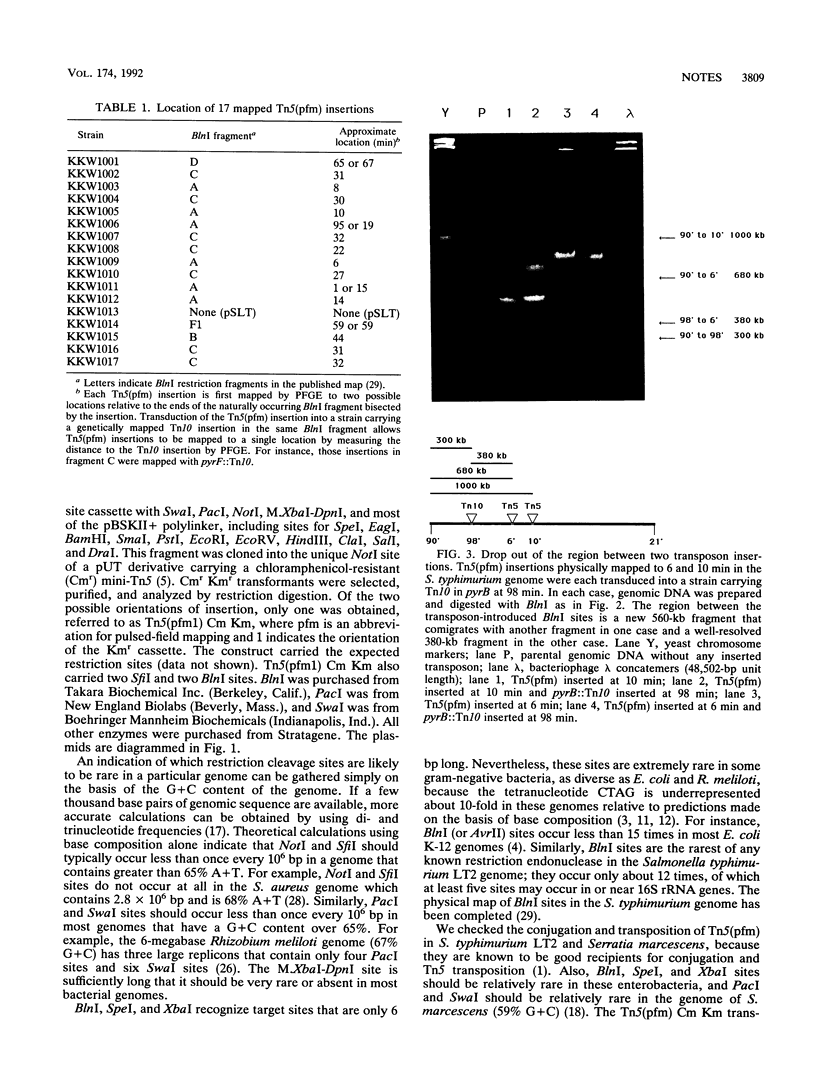
A polylinker with rare restriction sites was introduced into a mini-Tn5 derivative. These sites include M.XbaI-DpnI (TCTAGATCTAGA), which is rare in most bacterial genomes, SwaI (ATTTAAAT) and PacI (TTAATTAA), which are rare in G+C-rich genomes, NotI (GCGGCCGC) and SfiI (GGCCN5GGCC), which are rare in A+T-rich genomes, and BlnI (CCTAGG), SpeI (ACTAGT), and XbaI (TCTAGA), which are rare in the genomes of many gram-negative bacteria. This Tn5(pfm) (pulsed-field mapping) transposon carries resistance to chloramphenicol and kanamycin to allow selection in a wide variety of background genomes. This Tn5(pfm) was integrated randomly into the Salmonella typhimurium and Serratia marcescens genomes. Integration of the new rare SwaI, PacI, BlnI, SpeI, and XbaI sites was assayed by restriction digestion and pulsed-field gel electrophoresis. Tn5(pfm) constructs could be valuable tools for pulsed-field mapping of gram-negative bacterial genomes by assisting in the production of physical maps and restriction fragment catalogs. For the first applications of a Tn5(pfm), we bisected five of the six largest BlnI fragments in the S. typhimurium genome, bisected the linearized 90-kb pSLT plasmid, and used Tn5(pfm) and Tn10 to trisect the largest BlnI fragment.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Daniels D. L. The complete AvrII restriction map of the Escherichia coli genome and comparisons of several laboratory strains. Nucleic Acids Res. 1990 May 11;18(9):2649–2651. doi: 10.1093/nar/18.9.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellay R., Krisch H. M., Prentki P., Frey J. Omegon-Km: a transposable element designed for in vivo insertional mutagenesis and cloning of genes in gram-negative bacteria. Gene. 1989;76(2):215–226. doi: 10.1016/0378-1119(89)90162-5. [DOI] [PubMed] [Google Scholar]
- Gardiner K., Laas W., Patterson D. Fractionation of large mammalian DNA restriction fragments using vertical pulsed-field gradient gel electrophoresis. Somat Cell Mol Genet. 1986 Mar;12(2):185–195. doi: 10.1007/BF01560665. [DOI] [PubMed] [Google Scholar]
- Hanish J., McClelland M. Activity of DNA modification and restriction enzymes in KGB, a potassium glutamate buffer. Gene Anal Tech. 1988 Sep-Oct;5(5):105–107. doi: 10.1016/0735-0651(88)90005-2. [DOI] [PubMed] [Google Scholar]
- Hanish J., McClelland M. Enzymatic cleavage of a bacterial chromosome at a transposon-inserted rare site. Nucleic Acids Res. 1991 Feb 25;19(4):829–832. doi: 10.1093/nar/19.4.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob M., Grimes E., Szybalski W. Conferring operator specificity on restriction endonucleases. Science. 1988 Aug 26;241(4869):1084–1086. doi: 10.1126/science.2842862. [DOI] [PubMed] [Google Scholar]
- McClelland M., Bhagwat A. S. Biased DNA repair. Nature. 1992 Feb 13;355(6361):595–596. doi: 10.1038/355595b0. [DOI] [PubMed] [Google Scholar]
- McClelland M., Jones R., Patel Y., Nelson M. Restriction endonucleases for pulsed field mapping of bacterial genomes. Nucleic Acids Res. 1987 Aug 11;15(15):5985–6005. doi: 10.1093/nar/15.15.5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M., Nelson M. Enhancement of the apparent cleavage specificities of restriction endonucleases: applications to megabase mapping of chromosomes. Gene Amplif Anal. 1987;5:257–282. [PubMed] [Google Scholar]
- Michiels T., Popoff M. Y., Durviaux S., Coynault C., Cornelis G. A new method for the physical and genetic mapping of large plasmids: application to the localisation of the virulence determinants on the 90 kb plasmid of Salmonella typhimurium. Microb Pathog. 1987 Aug;3(2):109–116. doi: 10.1016/0882-4010(87)90069-6. [DOI] [PubMed] [Google Scholar]
- Miller V. L., Mekalanos J. J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988 Jun;170(6):2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteilhet C., Perrin A., Thierry A., Colleaux L., Dujon B. Purification and characterization of the in vitro activity of I-Sce I, a novel and highly specific endonuclease encoded by a group I intron. Nucleic Acids Res. 1990 Mar 25;18(6):1407–1413. doi: 10.1093/nar/18.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka A., Sugisaki H., Takanami M. Nucleotide sequence of the kanamycin resistance transposon Tn903. J Mol Biol. 1981 Apr 5;147(2):217–226. doi: 10.1016/0022-2836(81)90438-1. [DOI] [PubMed] [Google Scholar]
- Poddar S. K., McClelland M. Restriction fragment fingerprint and genome sizes of Staphylococcus species using pulsed-field gel electrophoresis and infrequent cleaving enzymes. DNA Cell Biol. 1991 Nov;10(9):663–669. doi: 10.1089/dna.1991.10.663. [DOI] [PubMed] [Google Scholar]
- Schwartz D. C., Cantor C. R. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984 May;37(1):67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- Smith C. L., Econome J. G., Schutt A., Klco S., Cantor C. R. A physical map of the Escherichia coli K12 genome. Science. 1987 Jun 12;236(4807):1448–1453. doi: 10.1126/science.3296194. [DOI] [PubMed] [Google Scholar]
- Smith C. L., Kolodner R. D. Mapping of Escherichia coli chromosomal Tn5 and F insertions by pulsed field gel electrophoresis. Genetics. 1988 Jun;119(2):227–236. doi: 10.1093/genetics/119.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobral B. W., Honeycutt R. J., Atherly A. G., McClelland M. Electrophoretic separation of the three Rhizobium meliloti replicons. J Bacteriol. 1991 Aug;173(16):5173–5180. doi: 10.1128/jb.173.16.5173-5180.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel S. A., Dervan P. B. Single-site enzymatic cleavage of yeast genomic DNA mediated by triple helix formation. Nature. 1991 Mar 14;350(6314):172–174. doi: 10.1038/350172a0. [DOI] [PubMed] [Google Scholar]
- Weil M. D., McClelland M. Enzymatic cleavage of a bacterial genome at a 10-base-pair recognition site. Proc Natl Acad Sci U S A. 1989 Jan;86(1):51–55. doi: 10.1073/pnas.86.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K. K., McClelland M. A BlnI restriction map of the Salmonella typhimurium LT2 genome. J Bacteriol. 1992 Mar;174(5):1656–1661. doi: 10.1128/jb.174.5.1656-1661.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lorenzo V., Herrero M., Jakubzik U., Timmis K. N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990 Nov;172(11):6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]