Abstract
Cultivated maize (Zea mays) and several other members of the Tribe Andropogoneae produce unisexual florets. In maize, the formation of two staminate florets in each spikelet on the tassel and a single pistillate floret in each spikelet on the ear includes a pistil abortion process that requires the action of the TASSELSEED2 gene. In Eastern gamagrass (Tripsacum dactyloides) the GYNOMONOECIOUS SEX FORM1 gene appears to perform a similar role in pistil abortion. These genes were shown to be homeologs by restriction fragment length polymorphism mapping and by the failure of the gsf1 and ts2 alleles to complement one another in intergeneric hybrids. Molecular analysis of the gsf1 allele shows that it is caused by a 1.4-kb deletion mutation. Both TASSELSEED2 and GYNOMONOECIOUS SEX FORM1 show similar expression patterns in subepidermal cells of pistils just before abortion. These results suggest that the formation of staminate florets in the Andropogoneae represents a monophyletic trait.
Maize and Eastern gamagrass are two monoecious species in the tribe Andropogoneae of the family Poaceae. Several members of this tribe, including maize and gamagrass, produce unisexual flowers, called “florets” in grasses. In both maize and Eastern gamagrass, two florets, the upper and lower florets, enclosed by a pair of leaf-like glumes, form a spikelet (Fig. 1C), the unit of grass inflorescences. In maize, the male and female florets are segregated in separate inflorescences. Two staminate florets are formed in each spikelet of the terminal inflorescence, the tassel, and a solitary pistillate floret is formed in each spikelet of the axillary inflorescence, the ear. Both tassel and ear spikelets are paired (Fig. 1B; ref. 1). Gamagrass forms terminal and axillary inflorescences of mixed sexuality with the apical two–thirds to three–quarters comprised of paired staminate spikelets, each containing two staminate florets, and the basal portion bearing solitary pistillate spikelets containing only one functional pistillate floret (2). Pistillate spikelets are unpaired because one of each pair of spikelets arrests and aborts in the pistillate region of the inflorescence (Figs. 1A and 2A). There is usually an abrupt transition between the zone of solitary pistillate spikelets and that of paired staminate spikelets, but occasionally a solitary staminate spikelet or a pair of pistillate spikelets is found at the boundary between these two zones (3).
Figure 1.
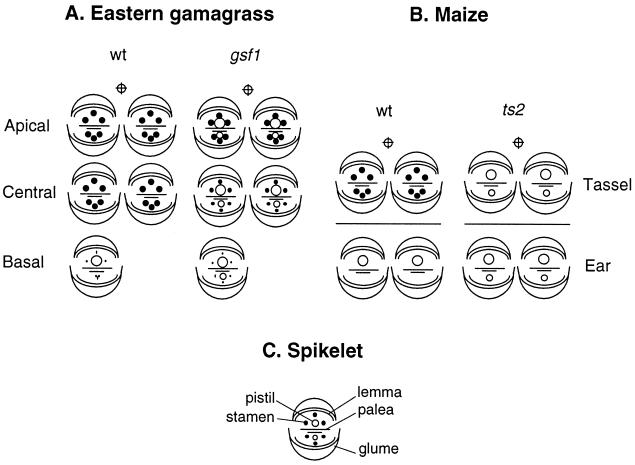
Diagrams of spikelet and floral structures of Eastern gamagrass and maize. (A) Floral diagrams of Gsf1 and gsf1 spikelets. Eastern gamagrass produces inflorescences with both staminate and pistillate spikelets. Staminate spikelets are paired and located on the upper two–thirds of the inflorescence. Each staminate spikelet contains two functional male florets. Pistillate spikelets are solitary, contain one functional pistil, and are located on the basal one–third of the inflorescence. In the gsf1 mutant, there is no pistil abortion. As a result, every floret contains a functional pistil. Stamens develop to different degrees, from rudimentary structures in the basal region of the inflorescence to fully developed ones at the apex. The ratio of solitary-to-paired spikelets is unaffected by the gsf1 mutation. (B) Floral diagrams of Ts2 and ts2 spikelets. Maize forms separate male and female inflorescences: tassel and ear. Staminate spikelets are paired, and each spikelet has two male florets. Pistillate spikelets also are paired, and only the upper floret in each spikelet develops. In the ts2 mutant, every spikelet contains two female florets, and each floret has a well developed pistil and no stamens. (C) Keys to floral structures. An immature bisexual spikelet of maize or Eastern gamagrass is comprised of two florets, the upper and the secondary lower florets, subtended by a pair of glumes. Each floret contains a lemma, a palea, three stamens, and a pistil. Lodicules are not shown.
Figure 2.
The inflorescences of Tripsacum. The wild-type (A) inflorescence consists of solitary pistillate spikelets at the base and paired staminate spikelets above. The gsf1 inflorescence (B) is feminized; all florets except those in the most apical region contain well developed pistils.
In both maize and Eastern gamagrass, the unisexuality of florets is established through a process of selective elimination or maturation of preformed floral organs in an initially bisexual immature floret (1, 3). In staminate spikelets, preformed pistil initials degenerate while stamen initials mature. In pistillate spikelets, the development of all stamen initials and the pistil initials of the lower florets is blocked, with only the pistils of the upper florets reaching maturity. The process of pistil abortion in maize requires the action of the tasselseed1 and tasselseed2 genes (4, 5). Mutations in these genes result in the failure to abort pistils, including the pistils in tassel spikelets and the lower floret pistils of ear spikelets (Fig. 1B; ref. 6). Ts2 has been cloned and shown to encode a short chain dehydrogenase expressed in subepidermal cells of pistils just before abortion (8). Little is known about the genetics of unisexuality in gamagrass except for a spontaneous recessive mutation called gynomonoecious sex form1 (gsf1) found in Kansas (2). The gsf1 mutation blocks pistil abortion in the apical staminate spikelets as well as the lower florets of the basal pistillate spikelets. Pistil development in these spikelets is associated with arrest of stamen initials. The stamens are arrested at different developmental stages, from rudimentary structures at the base to fully developed stamens at the upper portion of the inflorescence, as shown in Figs. 1A and 3. The gsf1 mutation does not affect the pattern of spikelet abortion; the basal portion of the inflorescence still contains solitary spikelets. As a result of feminization by gsf1, seed production in this variant is increased 10- to 25-fold (9), an important agronomic trait for the utility of Eastern gamagrass as a foraging crop.
Figure 3.
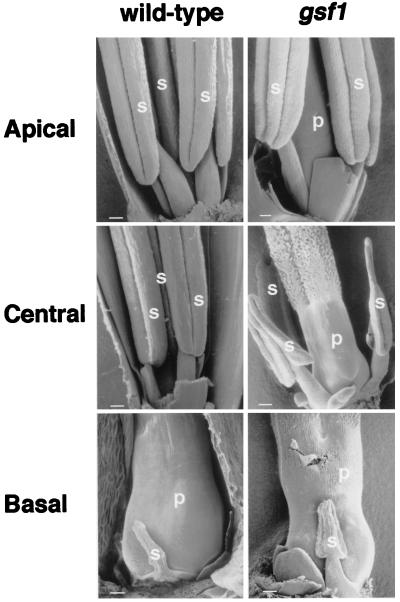
Scanning electron microscopy analysis of wild-type and gsf1 mutant florets. Staminate florets in the wild-type inflorescence (Left) contain three functional stamens and no remnants of pistils regardless of the location of florets; pistillate florets at the base each contain a fully developed pistil and three rudimentary stamens. In the gsf1 mutant (Right), all florets contain fully developed pistils. Stamens arrest at different developmental stages, from fully developed in the apical region, to immature in the central region, to rudimentary at the base. Wild-type (WW1218) and gsf1 mutant (WW1582) plants (2) were greenhouse grown. s, stamen; p, pistil. (Bar = 100 μm.)
MATERIALS AND METHODS
Plants and Intergeneric Crosses.
Maize ts2 stock was originally obtained from the Maize Genetics Cooperative, Champaign–Urbana, IL, and was grown in Hamden, CT for two generations. Tripsacum wild-type (WW1218), gsf1 mutant (WW1582), and heterozygous (WW1748) plants were obtained from the United States Department of Agriculture/Agricultural Research Service, Southern Plains Range Research Station, Woodward, OK. The intergeneric crosses were performed as described by Mangelsdorf and Reeves (10). The diploid (2n = 20), heterozygous (Ts2/ts2) maize plants were crossed as females by pollen from diploid (2n = 36), heterozygous (Gsf1/gsf1) Tripsacum plants (WW1748) (2). Hybrid seeds were harvested and kept at 4°C for more than 4 weeks. Afterward, hybrid seeds were surface-sterilized with 50% commercial bleach (Clorox, Oakland, CA) for 10 minutes and washed in distilled water, then germinated on Murashige minimal organics medium with 0.8% phytagar (GIBCO/BRL, Oakland, CA) at 28°C with a 16-h photoperiod. Two weeks after germination, seedlings were transferred to soil and grown in a greenhouse.
Scanning Electron Microscopy.
Samples for scanning electron microscopic analysis were prepared as described (8). Immature Tripsacum inflorescences were fixed in 50% ethanol/10% formaldehyde/5% acetic acid overnight, then dehydrated by passage through an ethanol series. Samples were then subjected to critical point dry and coated with gold/palladium. Coated inflorescences were examined in an International Scientific Instruments model SS40 scanning electron microscope.
Southern Blots and Sequence Analysis.
Southern analysis was carried out under stringent conditions as described (11). The gsf1 allele was cloned from mutant plants (WW1582), and the wild-type allele was cloned from a phenotyically normal plant found near the site that the first mutant plant was found in Ottawa County, Kansas (12). Both strands of wild-type and mutant alleles were sequenced.
RNA in Situ Hybridization.
Tripsacum and maize immature inflorescences (1–2 cm) were fixed in 4% formaldehyde in PBS solution. In situ hybridization was performed according to published methods (7). Sense and antisense riboprobes were derived from the maize Ts2 gene (8).
RESULTS
Mapping the gsf1 Trait.
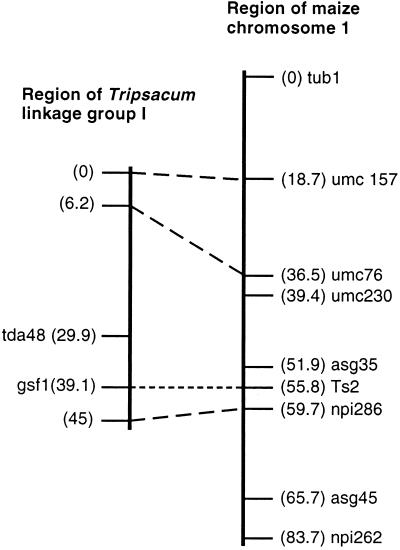
Eastern gamagrass Gsf1 and maize Ts2 genes have been suggested to be related genes based on phenotypic similarity (Fig. 2) and on linkage of each trait to molecular markers (13, 14). To investigate the possibility that Gsf1 and Ts2 are homeologous genes, we used Ts2 cDNA probes for Southern analysis of gamagrass F2 populations segregating for the gsf1 trait. Results (summarized in Fig. 4) indicated that gamagrass contained a single Ts2-hybridizing band that mapped to linkage group I, 5.9 cM from the maize-derived molecular marker npi286 and 9.2 cM from the gamagrass-derived molecular marker tda48. This position was closely linked (no recombinants detected in 113 individuals) to the gsf1 trait. The restriction fragment length polymorphism data also revealed that the Ts2 gene and gsf1 trait are located in the homeologous segments of maize chromosome 1 and gamagrass linkage group I, respectively (Fig. 4).
Figure 4.
The restriction fragment length polymorphism linkage maps of gamagrass linkage group I and maize chromosome 1. No recombination was observed between the Ts2 gene and the gsf1 phenotype in the Eastern gamagrass F2 mapping population (113 individuals) segregating 3:1 for the gsf1 trait (13, 14). Ts2 was mapped 3.9 cM distal to npi286 and 3.9 cM proximal to asg35 on chromosome 1 in the maize UMC IF2 population (54 individuals) (15). Comparative map analysis also revealed that two chromosomal regions (41 cM on maize chromosome 1 and 45 cM in gamagrass Group I) are homeologous segments. The order of the three “anchor loci,” npi286, umc76, and umc157, was conserved. Data were analyzed using mapmaker 2.0 (E. S. Lander, Whitehead Institute for Biomedical Research, Cambridge, MA). The molecular marker tda48 is derived from Eastern gamagrass; all others are derived from maize.
Cloning and Sequencing Gsf1 and gsf1 Alleles.
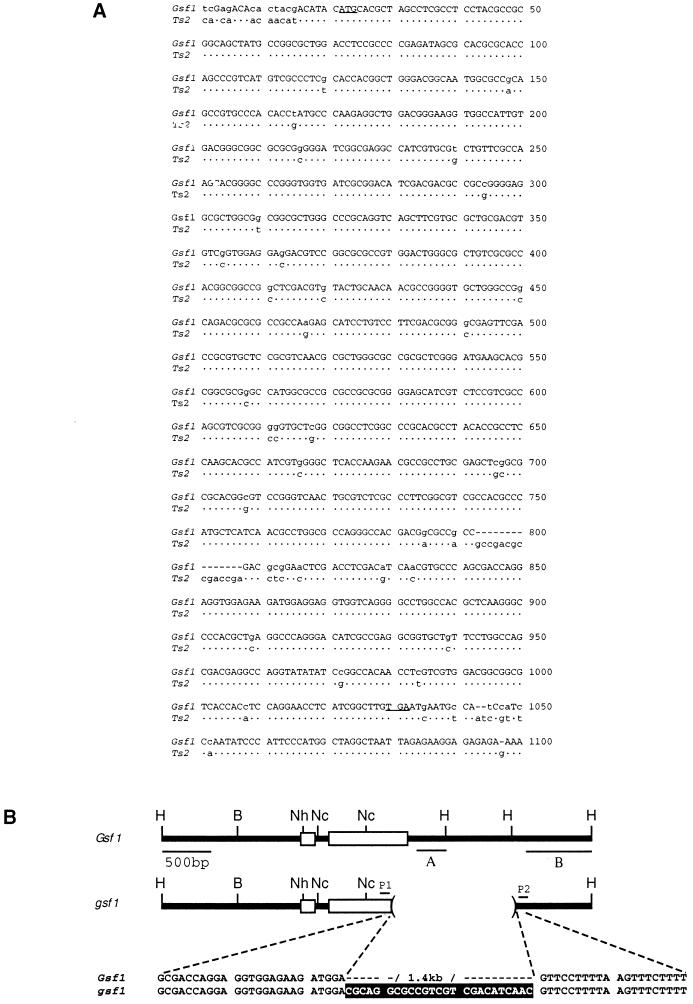
Further indication that the Gsf1 and Ts2 are homeologs came from molecular cloning of the gamagrass homology, which revealed that Gsf1 has a gene structure (two exons and one intron) similar to the maize Ts2 gene and that these two genes share greater than 90% identity in coding sequences (Fig. 5A). The predicted gamagrass protein shows 95% identity with the maize TS2 protein and contains three conserved motifs found in other short chain dehydrogenases: an N-terminal cofactor binding motif, a central catalytic motif, and a C-terminal domain of unknown significance (8, 16). Additional evidence that the gamagrass Ts2 homology represents the Gsf1 locus was obtained by comparing the Ts2 homologies in gsf1 and Gsf1 DNA. This analysis indicated that a major structural rearrangement occurred in the 3′ end of the coding region in gsf1 DNA. A restriction map generated by Southern data shows that the rearrangement was caused by a deletion of 1.4 kb (Fig. 5B). Sequence analysis of gsf1 DNA indicated that 25 bp of DNA of unknown origin was added between the breakpoints during deletion formation (Fig. 5B), a common phenomenon associated with deletions in maize (17). The deletion found in the gsf1 allele results in a predicted product containing a substitution of 53 amino acids for the last 163 nucleotides found in the Gsf1 allele. There were also minor sequence polymorphisms detected between the Ts2-homologous regions in Gsf1 and gsf1 DNA located in the noncoding regions (data not shown).
Figure 5.
Gsf1 and Ts2 genes. (A) Shown are the Gsf1 coding sequence (top line) with the start and stop codons underlined and the mismatches in the Ts2 gene sequence indicated below. The coding sequences from Gsf1 and Ts2 show more than 90% identity. (B) Results from Southern analysis and DNA sequencing of the gsf1 mutation are summarized. Probe A hybridized to Gsf1 DNA but not to gsf1 DNA. Probe B hybridized to both genomic DNAs, but the gsf1 fragment was 1.4 kb smaller than in wild type (data not shown). PCR analysis using P1 and P2 as primers also indicated that the gsf1 amplification product was 1.4 kb smaller than the product found in wild type. Sequence analysis of both alleles indicated that, between the deletion breakpoints, 25 bp of filler DNA is inserted (highlighted nucleotides). In addition to the 1.4-kb deletion, several minor nucleotide polymorphisms in intron and 5′ noncoding sequences were detected between wild-type and mutant alleles (not shown). H, HindIII; B, BamHI; Nh, NheI; Nc, NcoI.
We examined the expression of the gamagrass Ts2 homeolog in young staminate florets by RNA in situ hybridization. As seen with Ts2 expression in maize (8), the gamagrass gene also is expressed in the subepidermal cells of the pistil primordia before their abortion (Fig. 6).
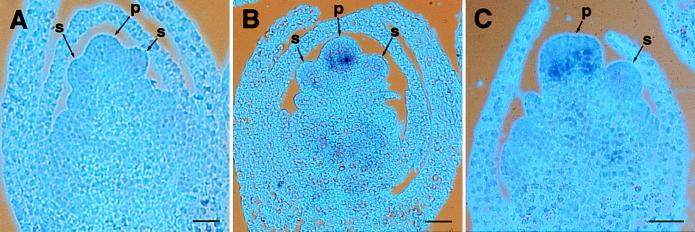
Figure 6.
The expression of Gsf1 gene detected by RNA in situ hybridization. Thin sections of Eastern gamagrass (A and B) and maize (C) immature staminate florets hybridized with maize Ts2 sense (A) and antisense (B and C) probes. The pistil (p) and stamen (s) primordia are indicated by arrows. (Bar = 50 μm.)
Complementation Test.
Complementation studies between Ts2 and Gsf1 were performed, and all four possible genotypic combinations of Gsf1 and Ts2 alleles among intergeneric F1 hybrids were identified by PCR analysis. Maize Ts2/ts2 plants were fertilized with pollen from gamagrass Gsf1/gsf1 plants, and 17 recovered hybrids flowered. Regardless of the genotype, all hybrids were highly tillered, and most shoots produced branched terminal inflorescences with a few solitary spikelets at the base and paired spikelets above. These phenotypes resembled those observed in other maize–gamagrass hybrids (18). Thirteen hybrids produced the expected wild-type pattern of solitary spikelets containing a single pistillate floret and paired spikelets containing two staminate florets (Fig. 7A). All 13 of these hybrids contained at least one copy of the functional gene Gsf1 or Ts2. Their genotypes were Gsf1/Ts2, Gsf1/ts2, or gsf1/Ts2 (Table 1). Four hybrids produced atypical inflorescences that were almost completely feminized except for 5–10 spikelets in the most apical region (Fig. 7B). The genotypes of these hybrids were determined to be gsf1/ts2 (Table 1). All spikelets on these atypical inflorescences contained a pair of florets with well developed pistils, but the stamens were arrested at different developmental stages (not shown), similar to what is seen in the gamagrass gsf1 mutant inflorescence. Stamen arrest occurred when initials were rudimentary in basal spikelets, but with increasing degrees of development seen in more apical regions. These results clearly showed that mutations in Ts2 and Gsf1 failed to complement each other genetically in intergeneric F1 hybrids.
Figure 7.
The inflorescences of maize–gamagrass intergeneric hybrids. (A) Representative inflorescence from a plant carrying at least one wild-type gene (Ts2 or Gsf1). (B) The feminized inflorescence from a plant with the genotype gsf1/ts2.
Table 1.
The phenotypic characterization of maize–Tripsacum intergeneric hybrids
| Genotype | Gsf1/Ts2 | Gsf1/ts2 | gsf1/Ts2 | gsf1/ts2 |
|---|---|---|---|---|
| Plants flowered, n | 6 | 2 | 5 | 4 |
| Branches per inflorescence | 6–12 | 4–6 | 6–12 | 6–10 |
| Paired spikelets on the main branch (X2) | 21–32 | 27 | 25–40 | 30–39 |
| Solitary spikelets on main axis | 0–4 | 0 | 0–5 | 0–2 |
| Pistillate spikelets on the main axis | 0–4 | 0 | 0–3 | (27–34) × 2 |
Intergeneric hybrid plants were genotyped by PCR analysis; Ts2 and ts2 alleles were identified by a microsatellite repeat located 3 kb downstream of the Ts2 gene in maize (19). Gsf1 and gsf1 alleles were distinguished using primers P1 and P2 (shown in Fig. 5B) amplifying a 300-bp fragment from gsf1 mutant DNA and a 1.6-kb product from wild-type DNA. Thirteen plants (genotyped as Gsf1/Ts2, Gsf1/ts2, or gsf1/Ts2) produced normal inflorescences (Fig. 7A). Four plants (genotyped as gsf1/ts2) formed feminized inflorescences (Fig. 7B).
DISCUSSION
We conclude that Gsf1 and Ts2 are homeologous genes based on restriction fragment length polymorphism linkage between Ts2 homology and the Gsf1 trait, the detection of a structural rearrangement associated with the mutant gsf1 allele, and noncomplementation of the maize ts2 and gamagrass gsf1 mutant alleles in intergeneric hybrids. Both genes perform similar roles in these species in that they are responsible for the abortion of pistils in staminate spikelets and the abortion of the lower floret pistils in pistillate spikelets. The Gsf1 gene does not appear to have a role in the process of spikelet abortion; the ratio of solitary to paired spikelets is unaffected by the gsf1 mutation, even though sexual fate is profoundly affected, indicating that spikelet abortion and pistil abortion pathways are distinct.
There are major differences in spatial organization of the sexes in these two species. In Eastern gamagrass, sexes are contained within a single inflorescence whereas in maize the sexes are separated into different inflorescences: tassels and ears. Yet despite these major morphological differences, the sex determination process appears to function through a common mechanism of TASSELSEED2-induced pistil abortion. Even though the maize tassel and ear have been modified by continual artificial selection and cultivation, the same genetic pathway formed by natural selection appears to be in use. This suggests that the Ts2/Gsf1-based sex determination mechanism for pistil abortion is ancestral to both Zea and Tripsacum. This pathway may have an even more ancient origin; based on phylogenetic analysis, the formation of staminate florets appears to be a monophyletic trait in the Andropogoneae (20, 21), and our results support the view that a common genetic mechanism for pistil abortion exists in at least two genera in the tribe. This shared trait indicates that sex determination studies in maize will have important evolutionary and mechanistic implications for understanding the forces of natural selection that resulted in unisexual flowers.
Acknowledgments
We thank A. Calderon–Urrea for maize Ts2 gene expression control and for help in RNA in situ experiments, and we thank T. Nelson, M. Ronemus, and C. Ticknor for comments on the manuscript. This work was supported by a grant from the National Institutes of Health (GM38148) to S.L.D. D.L. was supported by a graduate fellowship from the Rockefeller Foundation.
ABBREVIATIONS
- Ts2 and ts2
the wild-type and mutant alleles of the maize tasselseed2 gene
- Gsf1 and gsf1
the wild-type and mutant alleles of the Tripsacum gynomonoecious sex form1 gene
Footnotes
References
- 1.Cheng P C, Greyson R I, Walden D B. Am J Bot. 1983;70:450–462. [Google Scholar]
- 2.Dewald C L, Burson B L, de Wet J M J, Harlan J R. Am J Bot. 1987;74:1055–1059. [Google Scholar]
- 3.Camara-Hernandez J, Gambino S. Beitr Biol Pflanzen. 1992;67:295–303. [Google Scholar]
- 4.Dellaporta S L, Calderon-Urrea A. Science. 1994;266:1501–1505. doi: 10.1126/science.7985019. [DOI] [PubMed] [Google Scholar]
- 5.Irish E E, Langdale J A, Nelson T M. Dev Genet (Amsterdam) 1994;15:155–171. [Google Scholar]
- 6.Irish E E, Nelson T M. Am J Bot. 1993;80:292–299. [Google Scholar]
- 7.Schmidt R J, Veit B, Mandel M A, Mena M, Hake S, Yanofsky M F. Plant Cell. 1993;5:729–737. doi: 10.1105/tpc.5.7.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeLong A, Calderon-Urrea A, Dellaporta S L. Cell. 1993;74:757–768. doi: 10.1016/0092-8674(93)90522-r. [DOI] [PubMed] [Google Scholar]
- 9.Dewald C L, Dayton R S. Phytologia. 1985b;57:156. [Google Scholar]
- 10.Mangelsdorf P C, Reeves R G. J Hered. 1931;22:329–343. [Google Scholar]
- 11.Dellaporta S L, Moreno M A. In: Southern Blot Hybridization. Freeling M, Walbot V, editors. New York: Springer; 1993. pp. 569–572. [Google Scholar]
- 12.Dewald C L, Dayton R S. Crop Sci. 1985a;25:715. [Google Scholar]
- 13.Blakey C A, Coe E H, Dewald C L. Maize Genetics Cooperation Newsletter. 1994;68:35–36. [Google Scholar]
- 14.Blakey C A, Dewald C L, Coe E H. Genome. 1994;37:809–812. doi: 10.1139/g94-115. [DOI] [PubMed] [Google Scholar]
- 15.Gardiner J M, Coe E H, Melia-Hancock S, Hoisington D A, Chao S. Genetics. 1993;134:917–930. doi: 10.1093/genetics/134.3.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Persson B, Krook M, Jornvall H. Eur J Biochem. 1991;200:537–543. doi: 10.1111/j.1432-1033.1991.tb16215.x. [DOI] [PubMed] [Google Scholar]
- 17.Wessler S, Tarpley A, Purugganan M, Spell M, Okagaki R. Proc Natl Acad Sci USA. 1990;87:8731–8735. doi: 10.1073/pnas.87.22.8731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newell C A, deWet J M J. Am J Bot. 1974;61:45–53. [Google Scholar]
- 19.Calderon-Urrea A, Dellaporta S L. Maize Genetics Cooperation Newsletter. 1994;68:70. [Google Scholar]
- 20.Kellogg E A, Watson L. Bot Rev. 1993;59:273–321. [Google Scholar]
- 21.Kellogg E A, Birchler J A. Systematic Biol. 1993;42:415–439. [Google Scholar]