Abstract
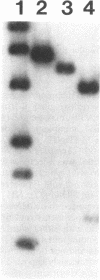
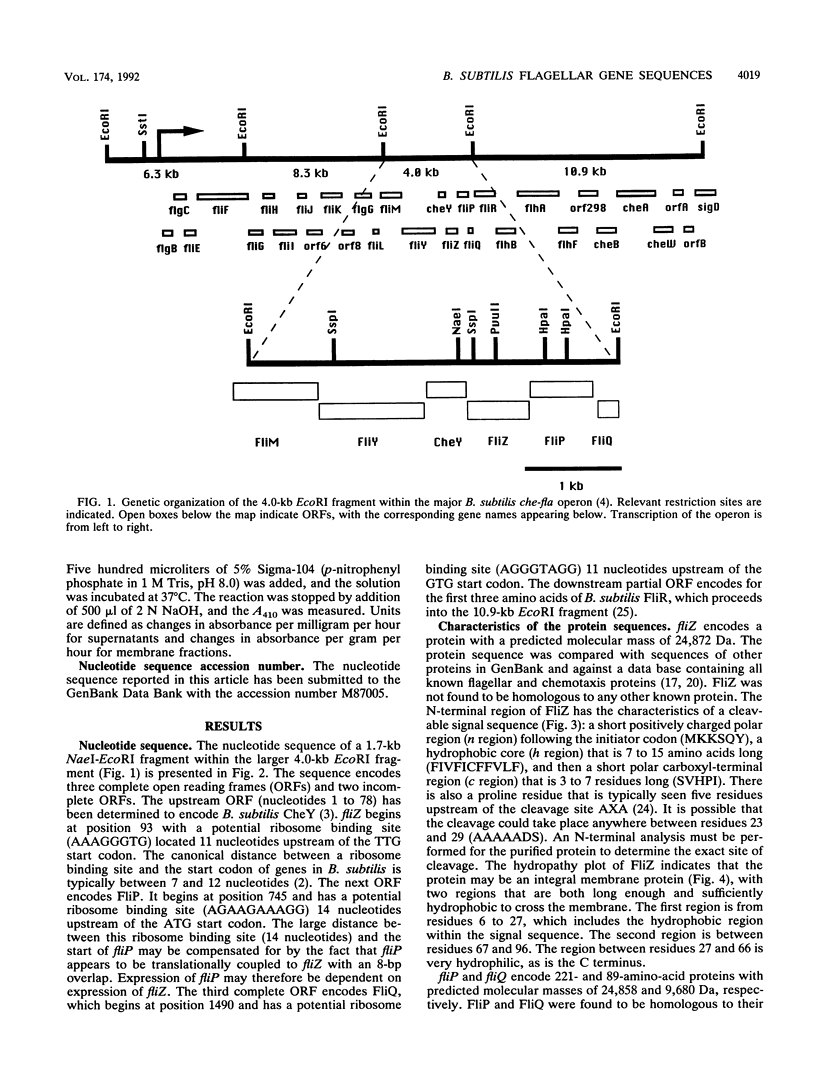
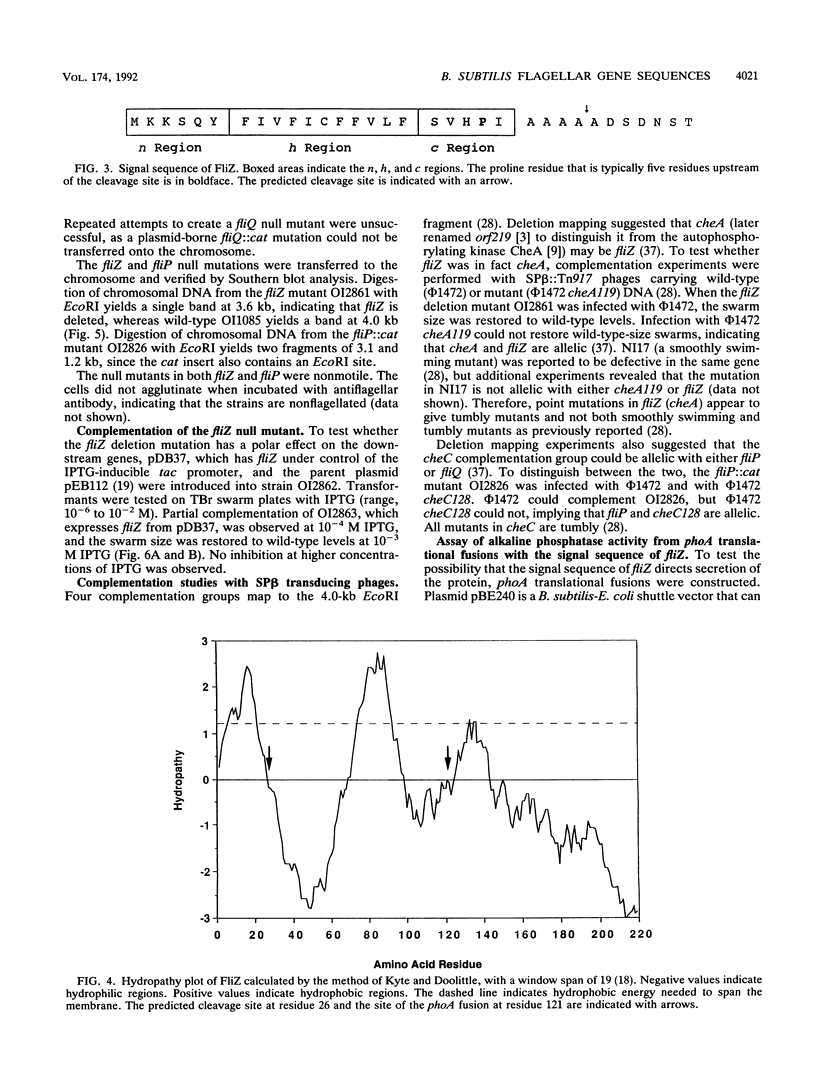
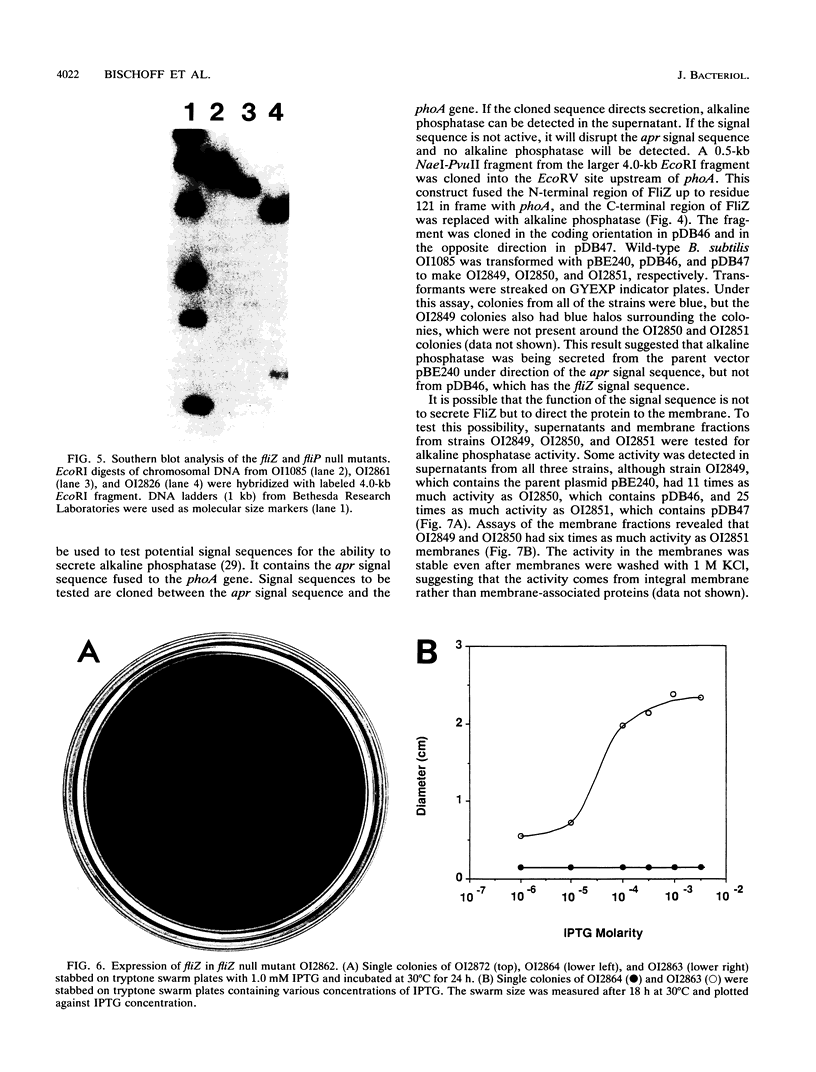
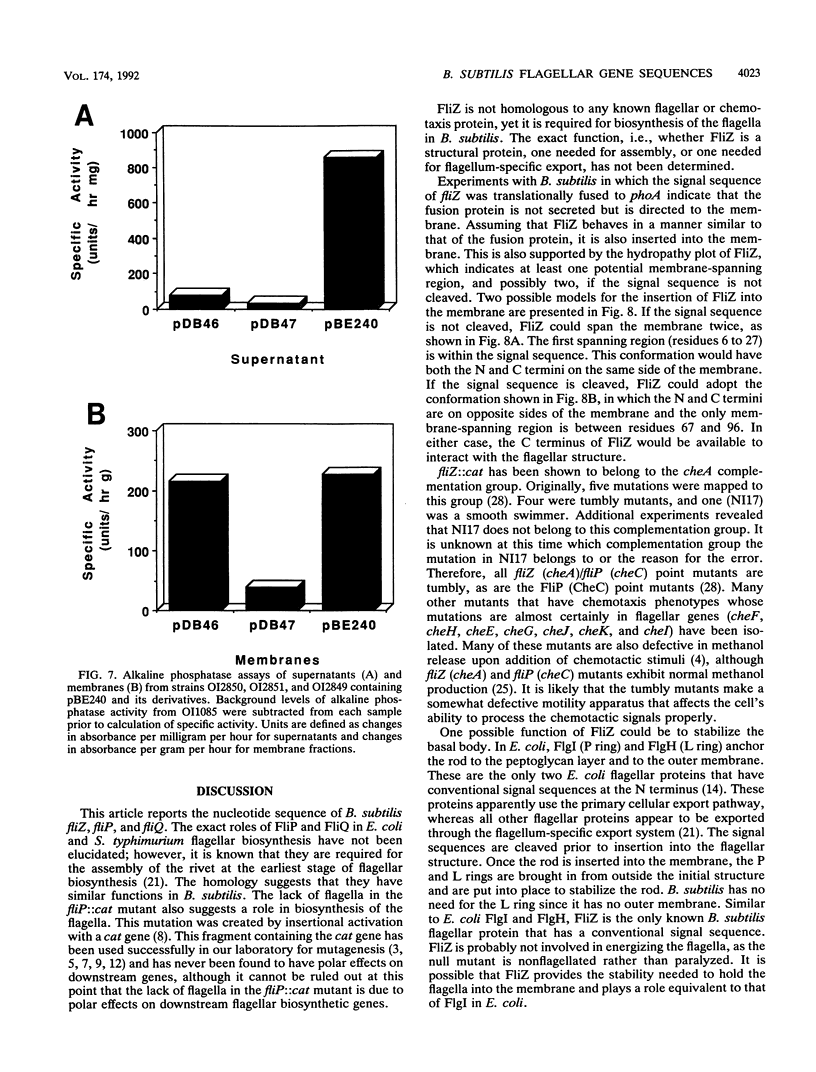
Three genes from the Bacillus subtilis major che-fla operon have been cloned and sequenced. Two of the genes encode proteins that are homologous to the Escherichia coli and Salmonella typhimurium flagellar biosynthetic proteins FliP and FliQ. The third gene, designated fliZ, encodes a 219-amino-acid protein with a predicted molecular mass of 24,872 Da. FliZ is not significantly homologous to any known proteins. Null mutants in fliP and fliZ do not have flagella; however, motility can be restored to the fliZ null mutant by expression of fliZ from a plasmid. FliZ has a conventional N-terminal signal sequence that does not direct secretion of the protein but appears to target the protein to the membrane. Two possible models of insertion of FliZ into the membrane are described.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albertini A. M., Caramori T., Crabb W. D., Scoffone F., Galizzi A. The flaA locus of Bacillus subtilis is part of a large operon coding for flagellar structures, motility functions, and an ATPase-like polypeptide. J Bacteriol. 1991 Jun;173(11):3573–3579. doi: 10.1128/jb.173.11.3573-3579.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Band L., Henner D. J. Bacillus subtilis requires a "stringent" Shine-Dalgarno region for gene expression. DNA. 1984;3(1):17–21. doi: 10.1089/dna.1.1984.3.17. [DOI] [PubMed] [Google Scholar]
- Bischoff D. S., Ordal G. W. Bacillus subtilis chemotaxis: a deviation from the Escherichia coli paradigm. Mol Microbiol. 1992 Jan;6(1):23–28. doi: 10.1111/j.1365-2958.1992.tb00833.x. [DOI] [PubMed] [Google Scholar]
- Bischoff D. S., Ordal G. W. Sequence and characterization of Bacillus subtilis CheB, a homolog of Escherichia coli CheY, and its role in a different mechanism of chemotaxis. J Biol Chem. 1991 Jul 5;266(19):12301–12305. [PubMed] [Google Scholar]
- Brickman E., Beckwith J. Analysis of the regulation of Escherichia coli alkaline phosphatase synthesis using deletions and phi80 transducing phages. J Mol Biol. 1975 Aug 5;96(2):307–316. doi: 10.1016/0022-2836(75)90350-2. [DOI] [PubMed] [Google Scholar]
- Covarrubias L., Bolivar F. Construction and characterization of new cloning vehicles. VI. Plasmid pBR329, a new derivative of pBR328 lacking the 482-base-pair inverted duplication. Gene. 1982 Jan;17(1):79–89. doi: 10.1016/0378-1119(82)90103-2. [DOI] [PubMed] [Google Scholar]
- Fuhrer D. K., Ordal G. W. Bacillus subtilis CheN, a homolog of CheA, the central regulator of chemotaxis in Escherichia coli. J Bacteriol. 1991 Dec;173(23):7443–7448. doi: 10.1128/jb.173.23.7443-7448.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman D. J., Ordal G. W. In vitro methylation and demethylation of methyl-accepting chemotaxis proteins in Bacillus subtilis. Biochemistry. 1984 Jun 5;23(12):2600–2606. doi: 10.1021/bi00307a010. [DOI] [PubMed] [Google Scholar]
- Hanlon D. W., Márquez-Magaña L. M., Carpenter P. B., Chamberlin M. J., Ordal G. W. Sequence and characterization of Bacillus subtilis CheW. J Biol Chem. 1992 Jun 15;267(17):12055–12060. [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Homma M., Komeda Y., Iino T., Macnab R. M. The flaFIX gene product of Salmonella typhimurium is a flagellar basal body component with a signal peptide for export. J Bacteriol. 1987 Apr;169(4):1493–1498. doi: 10.1128/jb.169.4.1493-1498.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iino T., Komeda Y., Kutsukake K., Macnab R. M., Matsumura P., Parkinson J. S., Simon M. I., Yamaguchi S. New unified nomenclature for the flagellar genes of Escherichia coli and Salmonella typhimurium. Microbiol Rev. 1988 Dec;52(4):533–535. doi: 10.1128/mr.52.4.533-535.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. J., Macnab R. M. Flagellar assembly in Salmonella typhimurium: analysis with temperature-sensitive mutants. J Bacteriol. 1990 Mar;172(3):1327–1339. doi: 10.1128/jb.172.3.1327-1339.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Leonhardt H., Alonso J. C. Construction of a shuttle vector for inducible gene expression in Escherichia coli and Bacillus subtilis. J Gen Microbiol. 1988 Mar;134(3):605–609. doi: 10.1099/00221287-134-3-605. [DOI] [PubMed] [Google Scholar]
- Macnab R. M., Parkinson J. S. Genetic analysis of the bacterial flagellum. Trends Genet. 1991 Jun;7(6):196–200. doi: 10.1016/0168-9525(91)90436-t. [DOI] [PubMed] [Google Scholar]
- Ordal G. W., Goldman D. J. Chemotaxis away from uncouplers of oxidative phosphorylation in Bacillus subtilis. Science. 1975 Sep 5;189(4205):802–805. doi: 10.1126/science.808854. [DOI] [PubMed] [Google Scholar]
- Ordal G. W., Nettleton D. O., Hoch J. A. Genetics of Bacillus subtilis chemotaxis: isolation and mapping of mutations and cloning of chemotaxis genes. J Bacteriol. 1983 Jun;154(3):1088–1097. doi: 10.1128/jb.154.3.1088-1097.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordal G. W., Parker H. M., Kirby J. R. Complementation and characterization of chemotaxis mutants of Bacillus subtilis. J Bacteriol. 1985 Nov;164(2):802–810. doi: 10.1128/jb.164.2.802-810.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne M. S., Jackson E. N. Use of alkaline phosphatase fusions to study protein secretion in Bacillus subtilis. J Bacteriol. 1991 Apr;173(7):2278–2282. doi: 10.1128/jb.173.7.2278-2282.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah A. H., Ordal G. W. In vivo and in vitro chemotactic methylation in Bacillus subtilis. J Bacteriol. 1981 Feb;145(2):958–965. doi: 10.1128/jb.145.2.958-965.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]
- Ying C. W., Scoffone F., Albertini A. M., Galizzi A., Ordal G. W. Properties of the Bacillus subtilis chemotaxis protein CheF, a homolog of the Salmonella typhimurium flagellar protein FliJ. J Bacteriol. 1991 Jun;173(11):3584–3586. doi: 10.1128/jb.173.11.3584-3586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuberi A. R., Bischoff D. S., Ordal G. W. Nucleotide sequence and characterization of a Bacillus subtilis gene encoding a flagellar switch protein. J Bacteriol. 1991 Jan;173(2):710–719. doi: 10.1128/jb.173.2.710-719.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuberi A. R., Ying C. W., Weinreich M. R., Ordal G. W. Transcriptional organization of a cloned chemotaxis locus of Bacillus subtilis. J Bacteriol. 1990 Apr;172(4):1870–1876. doi: 10.1128/jb.172.4.1870-1876.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuberi A. R., Ying C., Bischoff D. S., Ordal G. W. Gene-protein relationships in the flagellar hook-basal body complex of Bacillus subtilis: sequences of the flgB, flgC, flgG, fliE and fliF genes. Gene. 1991 May 15;101(1):23–31. doi: 10.1016/0378-1119(91)90220-6. [DOI] [PubMed] [Google Scholar]