Abstract
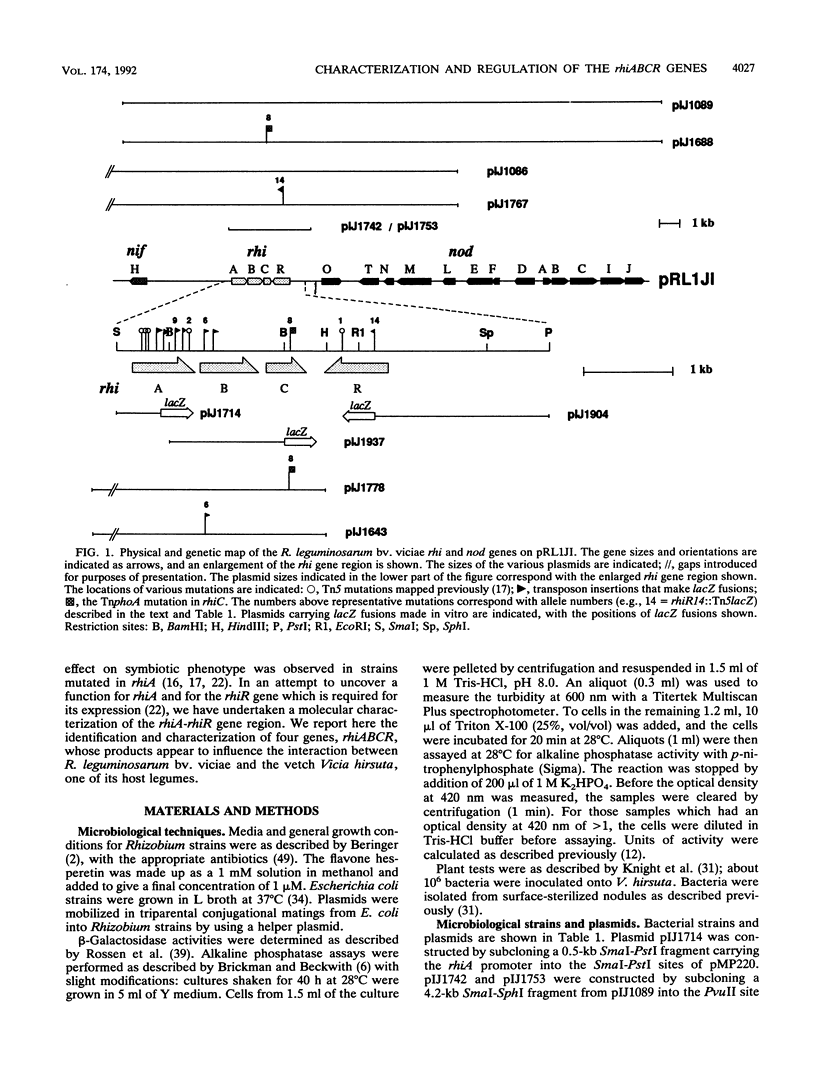
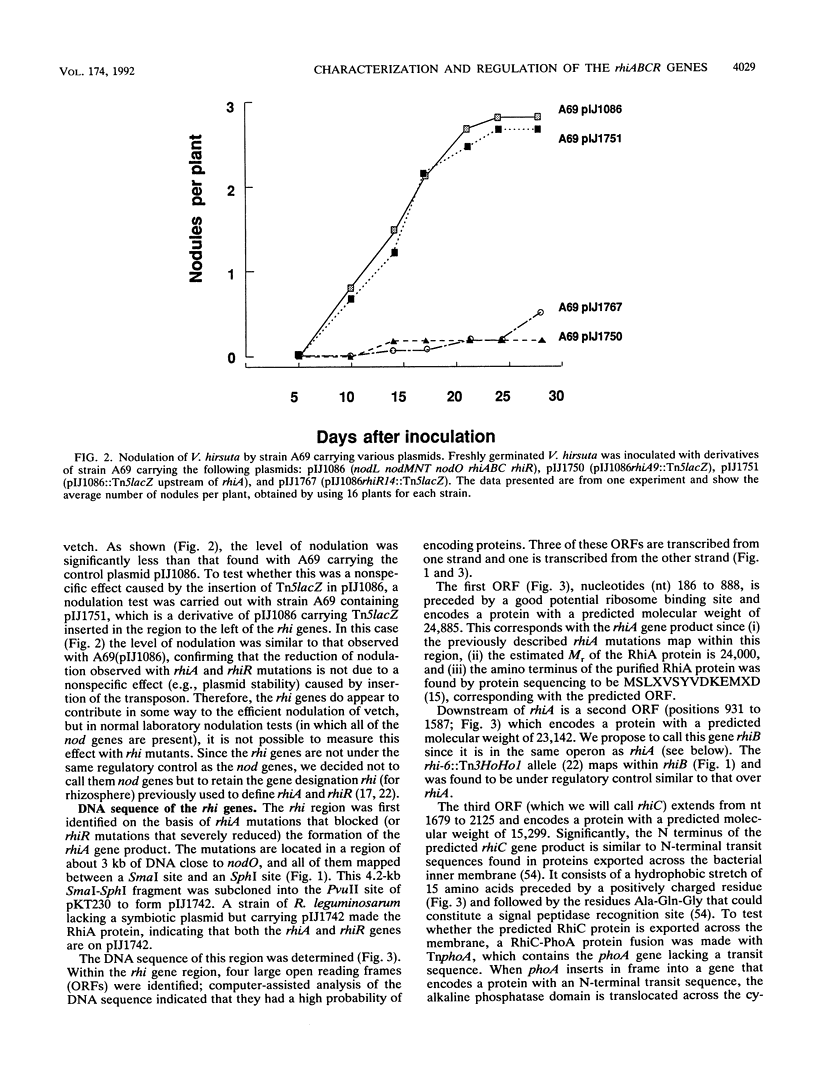
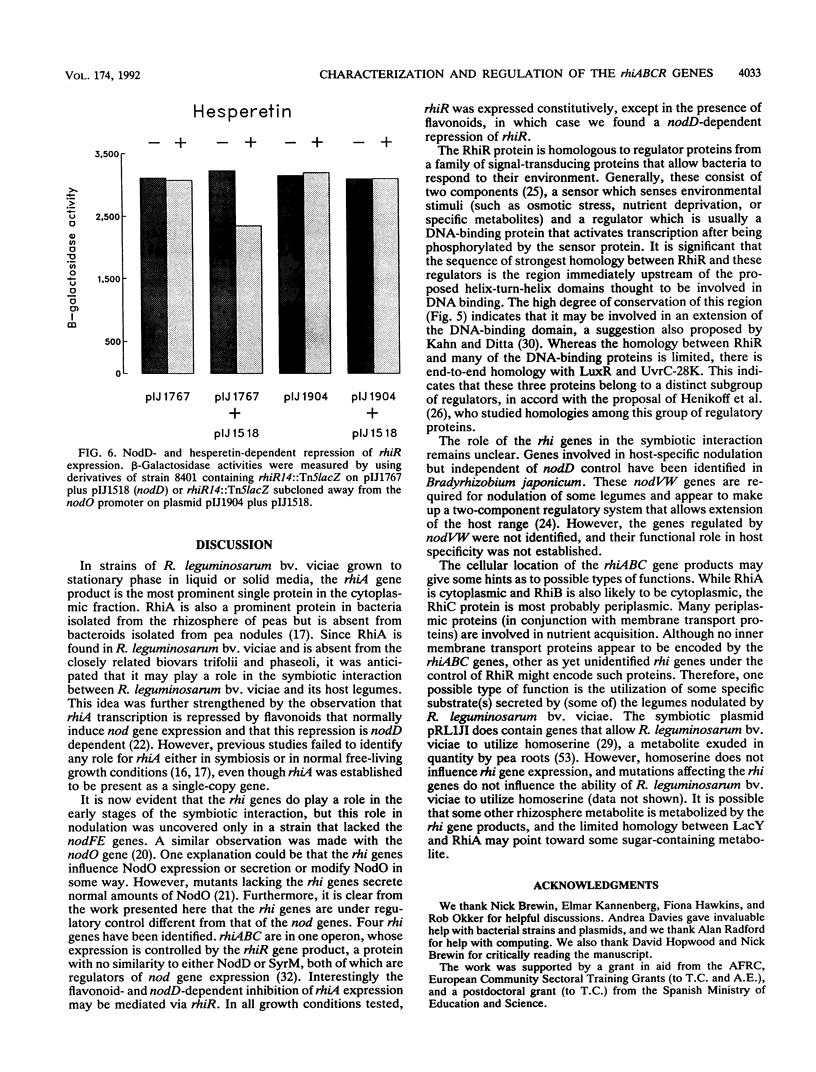
A group of four rhi (rhizosphere-expressed) genes from the symbiotic plasmid of Rhizobium leguminosarum biovar viciae has been characterized. Although mutation of the rhi genes does not normally affect nodulation, in the absence of the closely linked nodulation genes nodFEL, mutations in the rhi genes can influence the nodulation of the vetch Vicia hirsuta. The DNA sequence of the rhi gene region reveals four large open reading frames, three of them constituting an operon (rhiABC) transcribed convergently toward the fourth gene, rhiR. rhiABC are under the positive control of RhiR, the expression of which is repressed by flavonoids that normally induce nod gene expression. This repression, which requires the nodD gene product (the transcriptional activator of nod gene expression), may be due to a cis effect caused by a high level of NodD-dependent expression from the adjacent nodO promoter, which is transcribed divergently from rhiR. RhiR shows significant similarities to a subfamily of transcriptional regulators that includes the LuxR and UvrC-28K proteins. RhiA shows limited homology to a short domain of the lactose permease, LacY, close to a region thought to be involved in substrate binding. No strong homologies were found for the other rhi gene products. It appears that RhiA and RhiB are cytoplasmic, whereas RhiC is a periplasmic protein, since it has a typical N-terminal transit sequence and a rhiC-phoA protein fusion expresses alkaline phosphatase activity. The biochemical role of the rhi genes has not been established, but it appears that they may play a role in the plant-microbe interaction, possibly by allowing the bacteria to metabolize a plant-made metabolite.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aricó B., Miller J. F., Roy C., Stibitz S., Monack D., Falkow S., Gross R., Rappuoli R. Sequences required for expression of Bordetella pertussis virulence factors share homology with prokaryotic signal transduction proteins. Proc Natl Acad Sci U S A. 1989 Sep;86(17):6671–6675. doi: 10.1073/pnas.86.17.6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beringer J. E. R factor transfer in Rhizobium leguminosarum. J Gen Microbiol. 1974 Sep;84(1):188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- Boivin C., Barran L. R., Malpica C. A., Rosenberg C. Genetic analysis of a region of the Rhizobium meliloti pSym plasmid specifying catabolism of trigonelline, a secondary metabolite present in legumes. J Bacteriol. 1991 May;173(9):2809–2817. doi: 10.1128/jb.173.9.2809-2817.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickman E., Beckwith J. Analysis of the regulation of Escherichia coli alkaline phosphatase synthesis using deletions and phi80 transducing phages. J Mol Biol. 1975 Aug 5;96(2):307–316. doi: 10.1016/0022-2836(75)90350-2. [DOI] [PubMed] [Google Scholar]
- Cole S. T., Raibaud O. The nucleotide sequence of the malT gene encoding the positive regulator of the Escherichia coli maltose regulon. Gene. 1986;42(2):201–208. doi: 10.1016/0378-1119(86)90297-0. [DOI] [PubMed] [Google Scholar]
- Collins J. F., Coulson A. F., Lyall A. The significance of protein sequence similarities. Comput Appl Biosci. 1988 Mar;4(1):67–71. doi: 10.1093/bioinformatics/4.1.67. [DOI] [PubMed] [Google Scholar]
- Cubo M. T., Buendia-Claveria A. M., Beringer J. E., Ruiz-Sainz J. E. Melanin production by Rhizobium strains. Appl Environ Microbiol. 1988 Jul;54(7):1812–1817. doi: 10.1128/aem.54.7.1812-1817.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutting S., Mandelstam J. The nucleotide sequence and the transcription during sporulation of the gerE gene of Bacillus subtilis. J Gen Microbiol. 1986 Nov;132(11):3013–3024. doi: 10.1099/00221287-132-11-3013. [DOI] [PubMed] [Google Scholar]
- David M., Daveran M. L., Batut J., Dedieu A., Domergue O., Ghai J., Hertig C., Boistard P., Kahn D. Cascade regulation of nif gene expression in Rhizobium meliloti. Cell. 1988 Aug 26;54(5):671–683. doi: 10.1016/s0092-8674(88)80012-6. [DOI] [PubMed] [Google Scholar]
- Davis E. O., Johnston A. W. Analysis of three nodD genes in Rhizobium leguminosarum biovar phaseoli; nodD1 is preceded by noIE, a gene whose product is secreted from the cytoplasm. Mol Microbiol. 1990 Jun;4(6):921–932. doi: 10.1111/j.1365-2958.1990.tb00665.x. [DOI] [PubMed] [Google Scholar]
- Dibb N. J., Downie J. A., Brewin N. J. Identification of a rhizosphere protein encoded by the symbiotic plasmid of Rhizobium leguminosarum. J Bacteriol. 1984 May;158(2):621–627. doi: 10.1128/jb.158.2.621-627.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downie J. A., Ma Q. S., Knight C. D., Hombrecher G., Johnston A. W. Cloning of the symbiotic region of Rhizobium leguminosarum: the nodulation genes are between the nitrogenase genes and a nifA-like gene. EMBO J. 1983;2(6):947–952. doi: 10.1002/j.1460-2075.1983.tb01526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downie J. A., Surin B. P. Either of two nod gene loci can complement the nodulation defect of a nod deletion mutant of Rhizobium leguminosarum bv viciae. Mol Gen Genet. 1990 Jun;222(1):81–86. doi: 10.1007/BF00283027. [DOI] [PubMed] [Google Scholar]
- Economou A., Hamilton W. D., Johnston A. W., Downie J. A. The Rhizobium nodulation gene nodO encodes a Ca2(+)-binding protein that is exported without N-terminal cleavage and is homologous to haemolysin and related proteins. EMBO J. 1990 Feb;9(2):349–354. doi: 10.1002/j.1460-2075.1990.tb08117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economou A., Hawkins F. K., Downie J. A., Johnston A. W. Transcription of rhiA, a gene on a Rhizobium leguminosarum bv. viciae Sym plasmid, requires rhiR and is repressed by flavanoids that induce nod genes. Mol Microbiol. 1989 Jan;3(1):87–93. doi: 10.1111/j.1365-2958.1989.tb00107.x. [DOI] [PubMed] [Google Scholar]
- Friedrich M. J., Kadner R. J. Nucleotide sequence of the uhp region of Escherichia coli. J Bacteriol. 1987 Aug;169(8):3556–3563. doi: 10.1128/jb.169.8.3556-3563.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross R., Aricò B., Rappuoli R. Families of bacterial signal-transducing proteins. Mol Microbiol. 1989 Nov;3(11):1661–1667. doi: 10.1111/j.1365-2958.1989.tb00152.x. [DOI] [PubMed] [Google Scholar]
- Göttfert M., Grob P., Hennecke H. Proposed regulatory pathway encoded by the nodV and nodW genes, determinants of host specificity in Bradyrhizobium japonicum. Proc Natl Acad Sci U S A. 1990 Apr;87(7):2680–2684. doi: 10.1073/pnas.87.7.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Wallace J. C., Brown J. P. Finding protein similarities with nucleotide sequence databases. Methods Enzymol. 1990;183:111–132. doi: 10.1016/0076-6879(90)83009-x. [DOI] [PubMed] [Google Scholar]
- Henner D. J., Yang M., Ferrari E. Localization of Bacillus subtilis sacU(Hy) mutations to two linked genes with similarities to the conserved procaryotic family of two-component signalling systems. J Bacteriol. 1988 Nov;170(11):5102–5109. doi: 10.1128/jb.170.11.5102-5109.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn D., Ditta G. Modular structure of FixJ: homology of the transcriptional activator domain with the -35 binding domain of sigma factors. Mol Microbiol. 1991 Apr;5(4):987–997. doi: 10.1111/j.1365-2958.1991.tb00774.x. [DOI] [PubMed] [Google Scholar]
- Knight C. D., Rossen L., Robertson J. G., Wells B., Downie J. A. Nodulation inhibition by Rhizobium leguminosarum multicopy nodABC genes and analysis of early stages of plant infection. J Bacteriol. 1986 May;166(2):552–558. doi: 10.1128/jb.166.2.552-558.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondorosi A., Kondorosi E., John M., Schmidt J., Schell J. The role of nodulation genes in bacterium-plant communication. Genet Eng (N Y) 1991;13:115–136. doi: 10.1007/978-1-4615-3760-1_4. [DOI] [PubMed] [Google Scholar]
- Manoil C., Beckwith J. TnphoA: a transposon probe for protein export signals. Proc Natl Acad Sci U S A. 1985 Dec;82(23):8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy P. J., Heycke N., Trenz S. P., Ratet P., de Bruijn F. J., Schell J. Synthesis of an opine-like compound, a rhizopine, in alfalfa nodules is symbiotically regulated. Proc Natl Acad Sci U S A. 1988 Dec;85(23):9133–9137. doi: 10.1073/pnas.85.23.9133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossen L., Shearman C. A., Johnston A. W., Downie J. A. The nodD gene of Rhizobium leguminosarum is autoregulatory and in the presence of plant exudate induces the nodA,B,C genes. EMBO J. 1985 Dec 16;4(13A):3369–3373. doi: 10.1002/j.1460-2075.1985.tb04092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Shadel G. S., Baldwin T. O. The Vibrio fischeri LuxR protein is capable of bidirectional stimulation of transcription and both positive and negative regulation of the luxR gene. J Bacteriol. 1991 Jan;173(2):568–574. doi: 10.1128/jb.173.2.568-574.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S., Stark T. F., Beattie W. G., Moses R. E. Multiple control elements for the uvrC gene unit of Escherichia coli. Nucleic Acids Res. 1986 Mar 11;14(5):2301–2318. doi: 10.1093/nar/14.5.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaink H. P., Sheeley D. M., van Brussel A. A., Glushka J., York W. S., Tak T., Geiger O., Kennedy E. P., Reinhold V. N., Lugtenberg B. J. A novel highly unsaturated fatty acid moiety of lipo-oligosaccharide signals determines host specificity of Rhizobium. Nature. 1991 Nov 14;354(6349):125–130. doi: 10.1038/354125a0. [DOI] [PubMed] [Google Scholar]
- Staden R. An interactive graphics program for comparing and aligning nucleic acid and amino acid sequences. Nucleic Acids Res. 1982 May 11;10(9):2951–2961. doi: 10.1093/nar/10.9.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart V., Parales J., Jr, Merkel S. M. Structure of genes narL and narX of the nar (nitrate reductase) locus in Escherichia coli K-12. J Bacteriol. 1989 Apr;171(4):2229–2234. doi: 10.1128/jb.171.4.2229-2234.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout V., Gottesman S. RcsB and RcsC: a two-component regulator of capsule synthesis in Escherichia coli. J Bacteriol. 1990 Feb;172(2):659–669. doi: 10.1128/jb.172.2.659-669.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout V., Torres-Cabassa A., Maurizi M. R., Gutnick D., Gottesman S. RcsA, an unstable positive regulator of capsular polysaccharide synthesis. J Bacteriol. 1991 Mar;173(5):1738–1747. doi: 10.1128/jb.173.5.1738-1747.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surin B. P., Downie J. A. Characterization of the Rhizobium leguminosarum genes nodLMN involved in efficient host-specific nodulation. Mol Microbiol. 1988 Mar;2(2):173–183. doi: 10.1111/j.1365-2958.1988.tb00019.x. [DOI] [PubMed] [Google Scholar]
- Surin B. P., Watson J. M., Hamilton W. D., Economou A., Downie J. A. Molecular characterization of the nodulation gene, nodT, from two biovars of Rhizobium leguminosarum. Mol Microbiol. 1990 Feb;4(2):245–252. doi: 10.1111/j.1365-2958.1990.tb00591.x. [DOI] [PubMed] [Google Scholar]
- Trumble W. R., Viitanen P. V., Sarkar H. K., Poonian M. S., Kaback H. R. Site-directed mutagenesis of cys148 in the lac carrier protein of Escherichia coli. Biochem Biophys Res Commun. 1984 Mar 30;119(3):860–867. doi: 10.1016/0006-291x(84)90853-2. [DOI] [PubMed] [Google Scholar]
- Török I., Kondorosi E., Stepkowski T., Pósfai J., Kondorosi A. Nucleotide sequence of Rhizobium meliloti nodulation genes. Nucleic Acids Res. 1984 Dec 21;12(24):9509–9524. doi: 10.1093/nar/12.24.9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinrauch Y., Guillen N., Dubnau D. A. Sequence and transcription mapping of Bacillus subtilis competence genes comB and comA, one of which is related to a family of bacterial regulatory determinants. J Bacteriol. 1989 Oct;171(10):5362–5375. doi: 10.1128/jb.171.10.5362-5375.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Maagd R. A., Wijffelman C. A., Pees E., Lugtenberg B. J. Detection and subcellular localization of two Sym plasmid-dependent proteins of Rhizobium leguminosarum biovar viciae. J Bacteriol. 1988 Sep;170(9):4424–4427. doi: 10.1128/jb.170.9.4424-4427.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Maagd R. A., Wijfjes A. H., Spaink H. P., Ruiz-Sainz J. E., Wijffelman C. A., Okker R. J., Lugtenberg B. J. nodO, a new nod gene of the Rhizobium leguminosarum biovar viciae sym plasmid pRL1JI, encodes a secreted protein. J Bacteriol. 1989 Dec;171(12):6764–6770. doi: 10.1128/jb.171.12.6764-6770.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986 Jun 11;14(11):4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]