Abstract
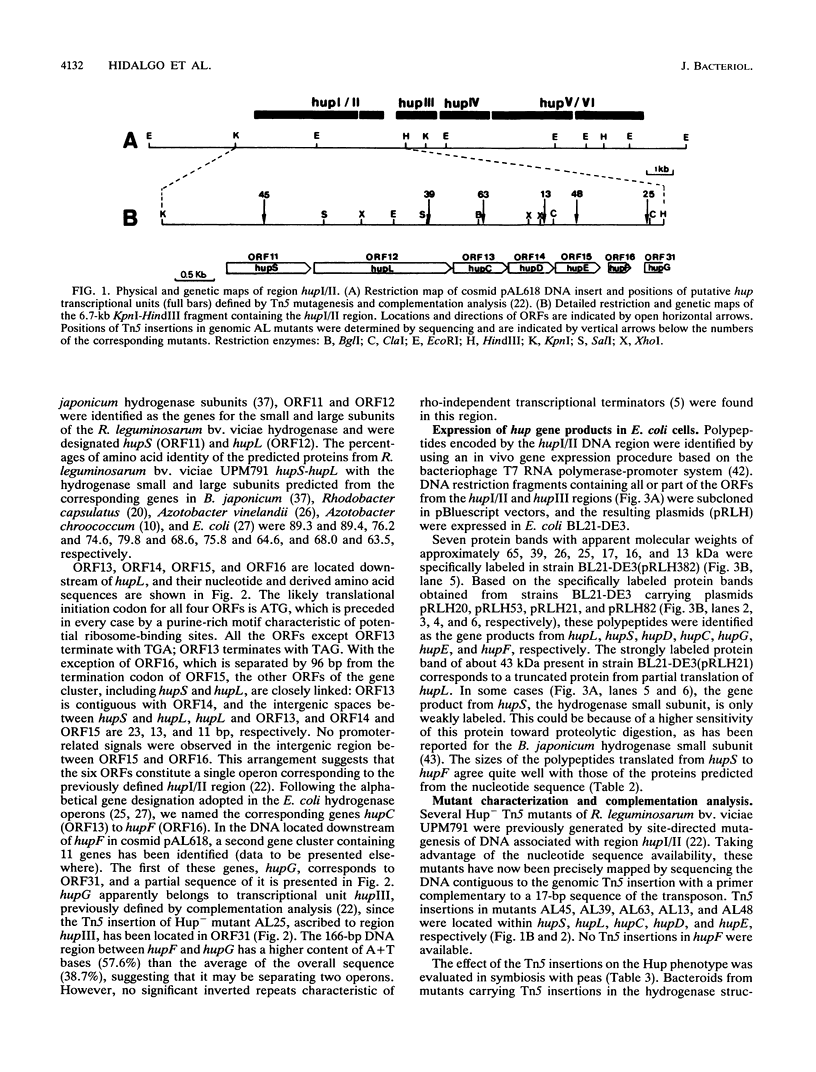
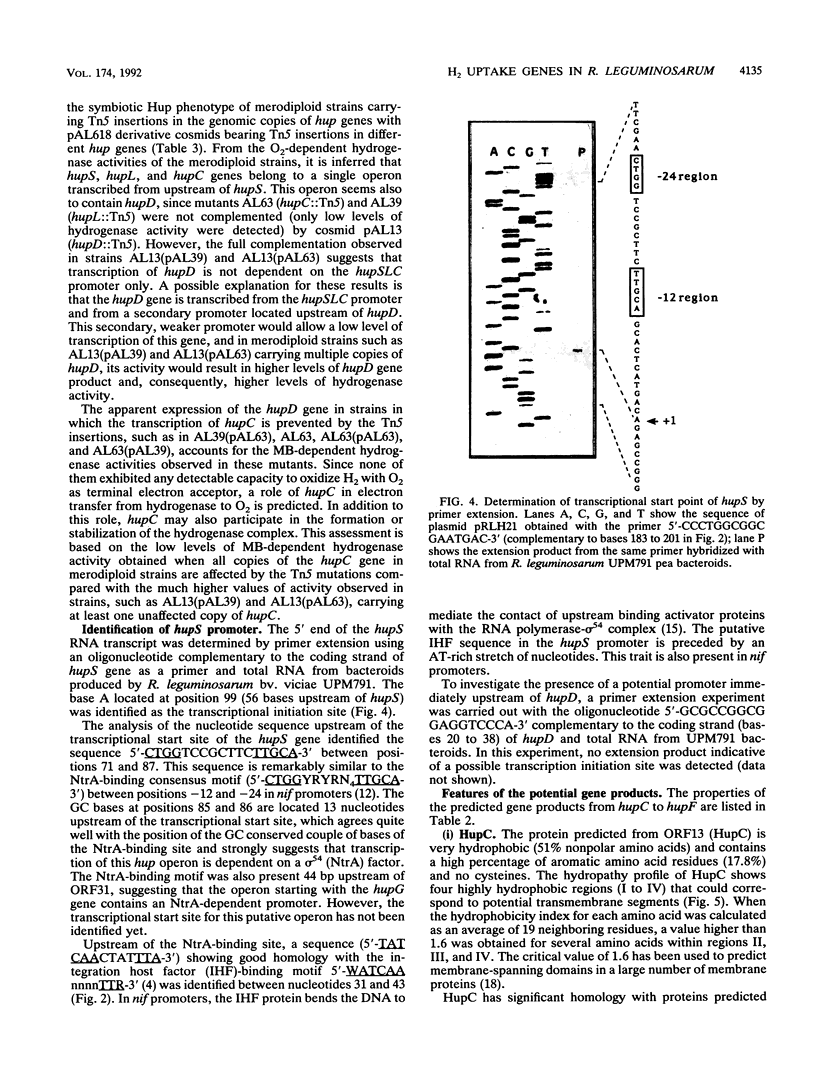
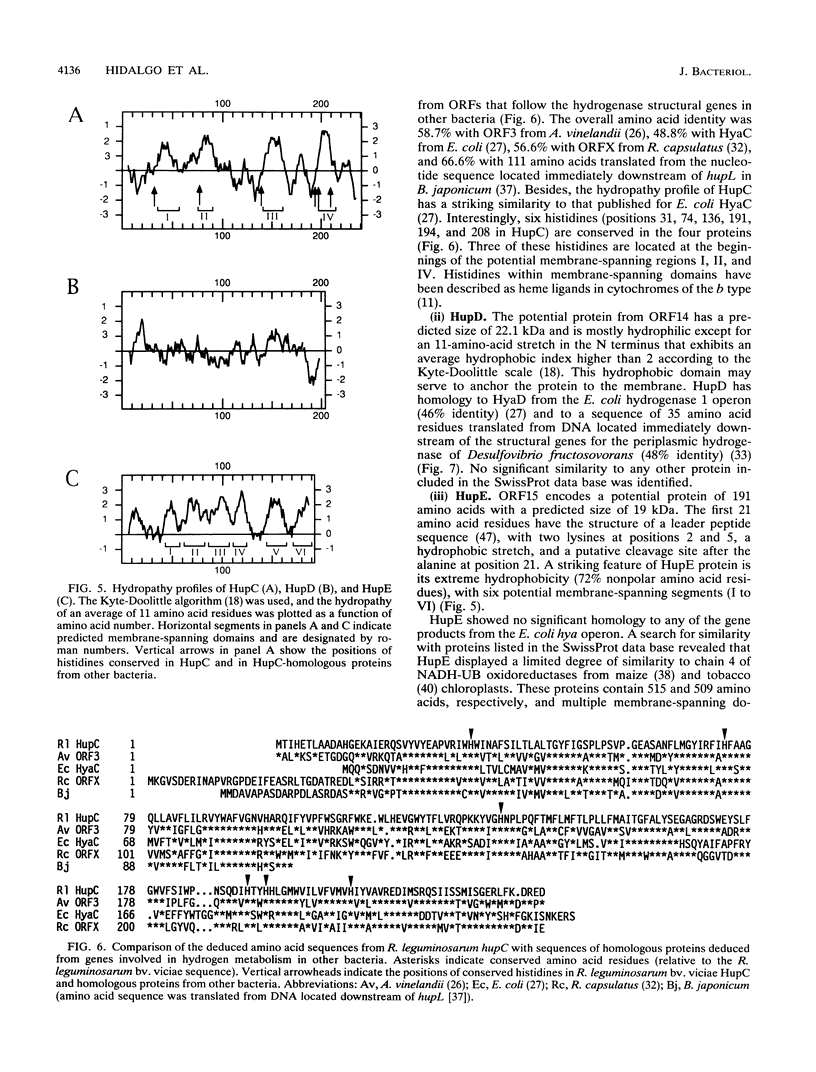
The nucleotide sequence of a 2.5-kbp region following the hydrogenase structural genes (hupSL) in the H2 uptake gene cluster from Rhizobium leguminosarum bv. viciae UPM791 was determined. Four closely linked genes encoding peptides of 27.9 (hupC), 22.1 (hupD), 19.0 (hupE), and 10.4 (hupF) kDa were identified immediately downstream of hupL. Proteins with comparable apparent molecular weights were detected by heterologous expression of these genes in Escherichia coli. The six genes, hupS to hupF, are arranged as an operon, and by mutant complementation analysis, it was shown that genes hupSLCD are cotranscribed. A transcription start site preceded by the -12 to -24 consensus sequence characteristic of NtrA-dependent promoters was identified upstream of hupS. On the basis of the lack of oxygen-dependent H2 uptake activity of a hupC::Tn5 mutant and on structural characteristics of the protein, we postulate that HupC is a b-type cytochrome involved in electron transfer from hydrogenase to oxygen. The product from hupE, which is needed for full hydrogenase activity, exhibited characteristics typical of a membrane protein. The features of HupC and HupE suggest that they form, together with the hydrogenase itself, a membrane-bound protein complex involved in hydrogen oxidation.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bairoch A. PROSITE: a dictionary of sites and patterns in proteins. Nucleic Acids Res. 1991 Apr 25;19 (Suppl):2241–2245. doi: 10.1093/nar/19.suppl.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhm R., Sauter M., Böck A. Nucleotide sequence and expression of an operon in Escherichia coli coding for formate hydrogenlyase components. Mol Microbiol. 1990 Feb;4(2):231–243. doi: 10.1111/j.1365-2958.1990.tb00590.x. [DOI] [PubMed] [Google Scholar]
- Cauvin B., Colbeau A., Vignais P. M. The hydrogenase structural operon in Rhodobacter capsulatus contains a third gene, hupM, necessary for the formation of a physiologically competent hydrogenase. Mol Microbiol. 1991 Oct;5(10):2519–2527. doi: 10.1111/j.1365-2958.1991.tb02098.x. [DOI] [PubMed] [Google Scholar]
- Craig N. L., Nash H. A. E. coli integration host factor binds to specific sites in DNA. Cell. 1984 Dec;39(3 Pt 2):707–716. doi: 10.1016/0092-8674(84)90478-1. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisbrenner G., Evans H. J. Carriers in electron transport from molecular hydrogen to oxygen in Rhizobium japonicum bacteroids. J Bacteriol. 1982 Mar;149(3):1005–1012. doi: 10.1128/jb.149.3.1005-1012.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisbrenner G., Evans H. J. Spectral evidence for a component involved in hydrogen metabolism of soybean nodule bacteroids. Plant Physiol. 1982 Dec;70(6):1667–1672. doi: 10.1104/pp.70.6.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans H. J., Harker A. R., Papen H., Russell S. A., Hanus F. J., Zuber M. Physiology, biochemistry, and genetics of the uptake hydrogenase in rhizobia. Annu Rev Microbiol. 1987;41:335–361. doi: 10.1146/annurev.mi.41.100187.002003. [DOI] [PubMed] [Google Scholar]
- Ford C. M., Garg N., Garg R. P., Tibelius K. H., Yates M. G., Arp D. J., Seefeldt L. C. The identification, characterization, sequencing and mutagenesis of the genes (hupSL) encoding the small and large subunits of the H2-uptake hydrogenase of Azotobacter chroococcum. Mol Microbiol. 1990 Jun;4(6):999–1008. doi: 10.1111/j.1365-2958.1990.tb00672.x. [DOI] [PubMed] [Google Scholar]
- Fridén H., Hederstedt L. Role of His residues in Bacillus subtilis cytochrome b558 for haem binding and assembly of succinate: quinone oxidoreductase (complex II). Mol Microbiol. 1990 Jun;4(6):1045–1056. doi: 10.1111/j.1365-2958.1990.tb00677.x. [DOI] [PubMed] [Google Scholar]
- Gussin G. N., Ronson C. W., Ausubel F. M. Regulation of nitrogen fixation genes. Annu Rev Genet. 1986;20:567–591. doi: 10.1146/annurev.ge.20.120186.003031. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III in DNA sequence analysis. Methods Enzymol. 1987;155:156–165. doi: 10.1016/0076-6879(87)55014-5. [DOI] [PubMed] [Google Scholar]
- Hidalgo E., Leyva A., Ruiz-Argüeso T. Nucleotide sequence of the hydrogenase structural genes from Rhizobium leguminosarum. Plant Mol Biol. 1990 Aug;15(2):367–370. doi: 10.1007/BF00036924. [DOI] [PubMed] [Google Scholar]
- Hoover T. R., Santero E., Porter S., Kustu S. The integration host factor stimulates interaction of RNA polymerase with NIFA, the transcriptional activator for nitrogen fixation operons. Cell. 1990 Oct 5;63(1):11–22. doi: 10.1016/0092-8674(90)90284-l. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leclerc M., Colbeau A., Cauvin B., Vignais P. M. Cloning and sequencing of the genes encoding the large and the small subunits of the H2 uptake hydrogenase (hup) of Rhodobacter capsulatus. Mol Gen Genet. 1988 Sep;214(1):97–107. doi: 10.1007/BF00340186. [DOI] [PubMed] [Google Scholar]
- Leyva A., Palacios J. M., Mozo T., Ruiz-Argüeso T. Cloning and characterization of hydrogen uptake genes from Rhizobium leguminosarum. J Bacteriol. 1987 Nov;169(11):4929–4934. doi: 10.1128/jb.169.11.4929-4934.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyva A., Palacios J. M., Murillo J., Ruiz-Argüeso T. Genetic organization of the hydrogen uptake (hup) cluster from Rhizobium leguminosarum. J Bacteriol. 1990 Mar;172(3):1647–1655. doi: 10.1128/jb.172.3.1647-1655.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz S., Böhm R., Beier A., Böck A. Characterization of divergent NtrA-dependent promoters in the anaerobically expressed gene cluster coding for hydrogenase 3 components of Escherichia coli. Mol Microbiol. 1990 Jan;4(1):13–20. doi: 10.1111/j.1365-2958.1990.tb02010.x. [DOI] [PubMed] [Google Scholar]
- Lutz S., Jacobi A., Schlensog V., Böhm R., Sawers G., Böck A. Molecular characterization of an operon (hyp) necessary for the activity of the three hydrogenase isoenzymes in Escherichia coli. Mol Microbiol. 1991 Jan;5(1):123–135. doi: 10.1111/j.1365-2958.1991.tb01833.x. [DOI] [PubMed] [Google Scholar]
- Menon A. L., Stults L. W., Robson R. L., Mortenson L. E. Cloning, sequencing and characterization of the [NiFe]hydrogenase-encoding structural genes (hoxK and hoxG) from Azotobacter vinelandii. Gene. 1990 Nov 30;96(1):67–74. doi: 10.1016/0378-1119(90)90342-o. [DOI] [PubMed] [Google Scholar]
- Menon N. K., Robbins J., Peck H. D., Jr, Chatelus C. Y., Choi E. S., Przybyla A. E. Cloning and sequencing of a putative Escherichia coli [NiFe] hydrogenase-1 operon containing six open reading frames. J Bacteriol. 1990 Apr;172(4):1969–1977. doi: 10.1128/jb.172.4.1969-1977.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brian M. R., Maier R. J. Expression of cytochrome o in hydrogen uptake constitutive mutants of Rhizobium japonicum. J Bacteriol. 1985 Feb;161(2):507–514. doi: 10.1128/jb.161.2.507-514.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brian M. R., Maier R. J. Hydrogen metabolism in Rhizobium: energetics, regulation, enzymology and genetics. Adv Microb Physiol. 1988;29:1–52. doi: 10.1016/s0065-2911(08)60345-8. [DOI] [PubMed] [Google Scholar]
- O'Brian M. R., Maier R. J. Role of ubiquinone in hydrogen-dependent electron transport in Rhizobium japonicum. J Bacteriol. 1985 Feb;161(2):775–777. doi: 10.1128/jb.161.2.775-777.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios J. M., Murillo J., Leyva A., Ditta G., Ruiz-Argüeso T. Differential expression of hydrogen uptake (hup) genes in vegetative and symbiotic cells of Rhizobium leguminosarum. Mol Gen Genet. 1990 May;221(3):363–370. doi: 10.1007/BF00259401. [DOI] [PubMed] [Google Scholar]
- Richaud P., Vignais P. M., Colbeau A., Uffen R. L., Cauvin B. Molecular biology studies of the uptake hydrogenase of Rhodobacter capsulatus and Rhodocyclus gelatinosus. FEMS Microbiol Rev. 1990 Dec;7(3-4):413–418. doi: 10.1016/0378-1097(90)90488-c. [DOI] [PubMed] [Google Scholar]
- Rousset M., Dermoun Z., Hatchikian C. E., Bélaich J. P. Cloning and sequencing of the locus encoding the large and small subunit genes of the periplasmic [NiFe]hydrogenase from Desulfovibrio fructosovorans. Gene. 1990 Sep 28;94(1):95–101. doi: 10.1016/0378-1119(90)90473-5. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayavedra-Soto L. A., Powell G. K., Evans H. J., Morris R. O. Nucleotide sequence of the genetic loci encoding subunits of Bradyrhizobium japonicum uptake hydrogenase. Proc Natl Acad Sci U S A. 1988 Nov;85(22):8395–8399. doi: 10.1073/pnas.85.22.8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C. G., Schmitt H. J., Schild C., Tichy H. V., Lotz W. DNA sequence encoding the two structural genes for the uptake hydrogenase of Rhizobium leguminosarum bv. viciae B10. Nucleic Acids Res. 1990 Sep 11;18(17):5285–5285. doi: 10.1093/nar/18.17.5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K., Ohme M., Tanaka M., Wakasugi T., Hayashida N., Matsubayashi T., Zaita N., Chunwongse J., Obokata J., Yamaguchi-Shinozaki K. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 1986 Sep;5(9):2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short J. M., Fernandez J. M., Sorge J. A., Huse W. D. Lambda ZAP: a bacteriophage lambda expression vector with in vivo excision properties. Nucleic Acids Res. 1988 Aug 11;16(15):7583–7600. doi: 10.1093/nar/16.15.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Stults L. W., Moshiri F., Maier R. J. Aerobic purification of hydrogenase from Rhizobium japonicum by affinity chromatography. J Bacteriol. 1986 Jun;166(3):795–800. doi: 10.1128/jb.166.3.795-800.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toussaint B., Bosc C., Richaud P., Colbeau A., Vignais P. M. A mutation in a Rhodobacter capsulatus gene encoding an integration host factor-like protein impairs in vivo hydrogenase expression. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10749–10753. doi: 10.1073/pnas.88.23.10749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran-Betcke A., Warnecke U., Böcker C., Zaborosch C., Friedrich B. Cloning and nucleotide sequences of the genes for the subunits of NAD-reducing hydrogenase of Alcaligenes eutrophus H16. J Bacteriol. 1990 Jun;172(6):2920–2929. doi: 10.1128/jb.172.6.2920-2929.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada K., Wada Y., Doi H., Ishibashi F., Gojobori T., Ikemura T. Codon usage tabulated from the GenBank genetic sequence data. Nucleic Acids Res. 1991 Apr 25;19 (Suppl):1981–1986. doi: 10.1093/nar/19.suppl.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widger W. R., Cramer W. A., Herrmann R. G., Trebst A. Sequence homology and structural similarity between cytochrome b of mitochondrial complex III and the chloroplast b6-f complex: position of the cytochrome b hemes in the membrane. Proc Natl Acad Sci U S A. 1984 Feb;81(3):674–678. doi: 10.1073/pnas.81.3.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Aubenton Carafa Y., Brody E., Thermes C. Prediction of rho-independent Escherichia coli transcription terminators. A statistical analysis of their RNA stem-loop structures. J Mol Biol. 1990 Dec 20;216(4):835–858. doi: 10.1016/s0022-2836(99)80005-9. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Signal sequences. The limits of variation. J Mol Biol. 1985 Jul 5;184(1):99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]