Abstract
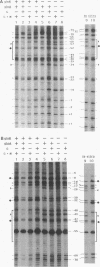
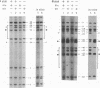
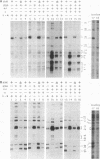
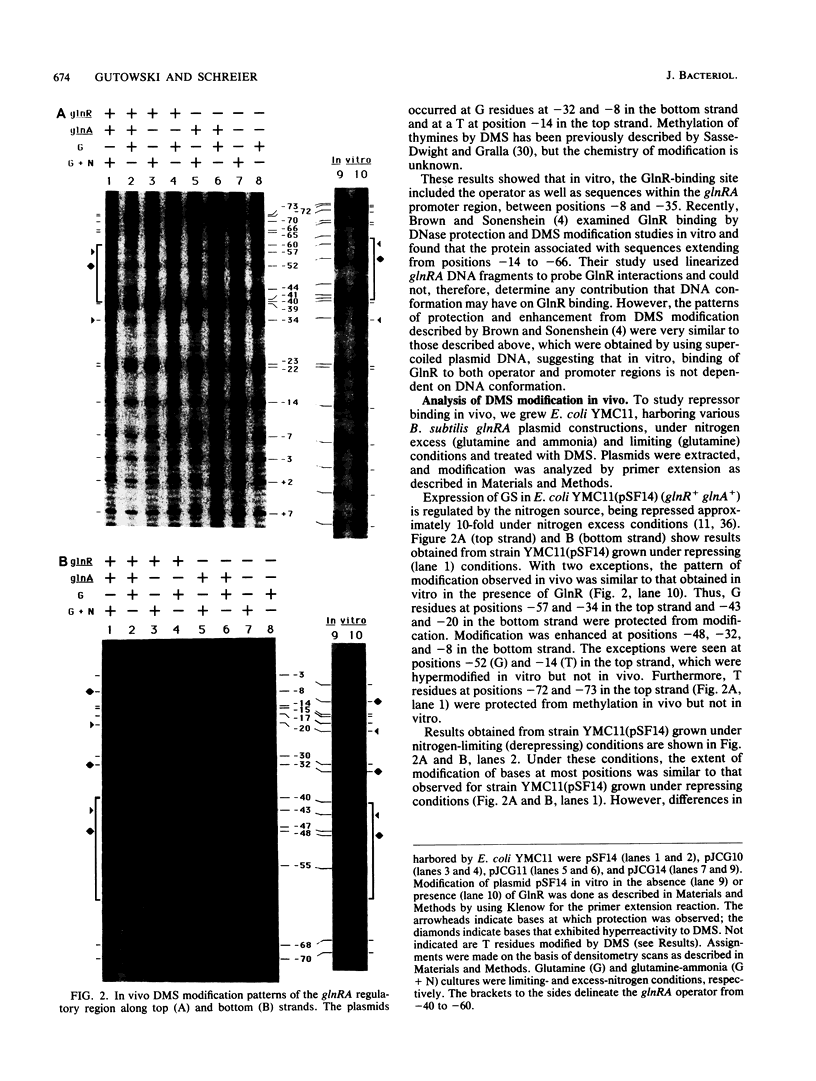
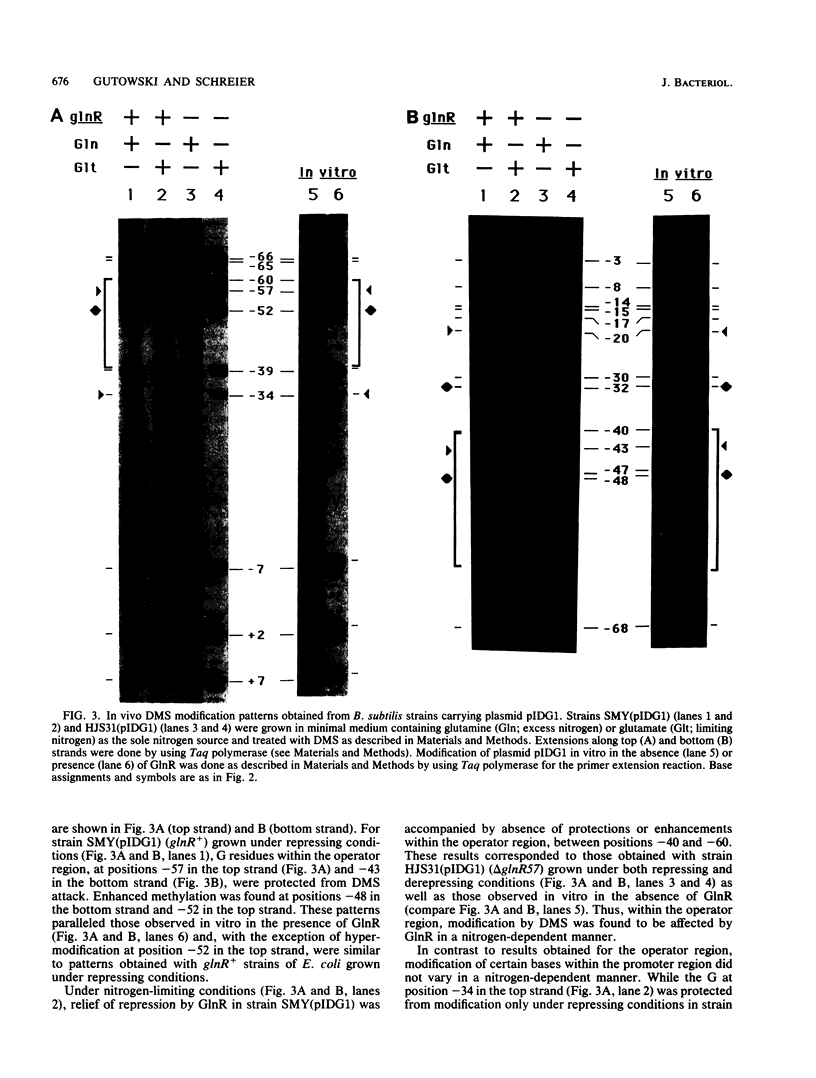
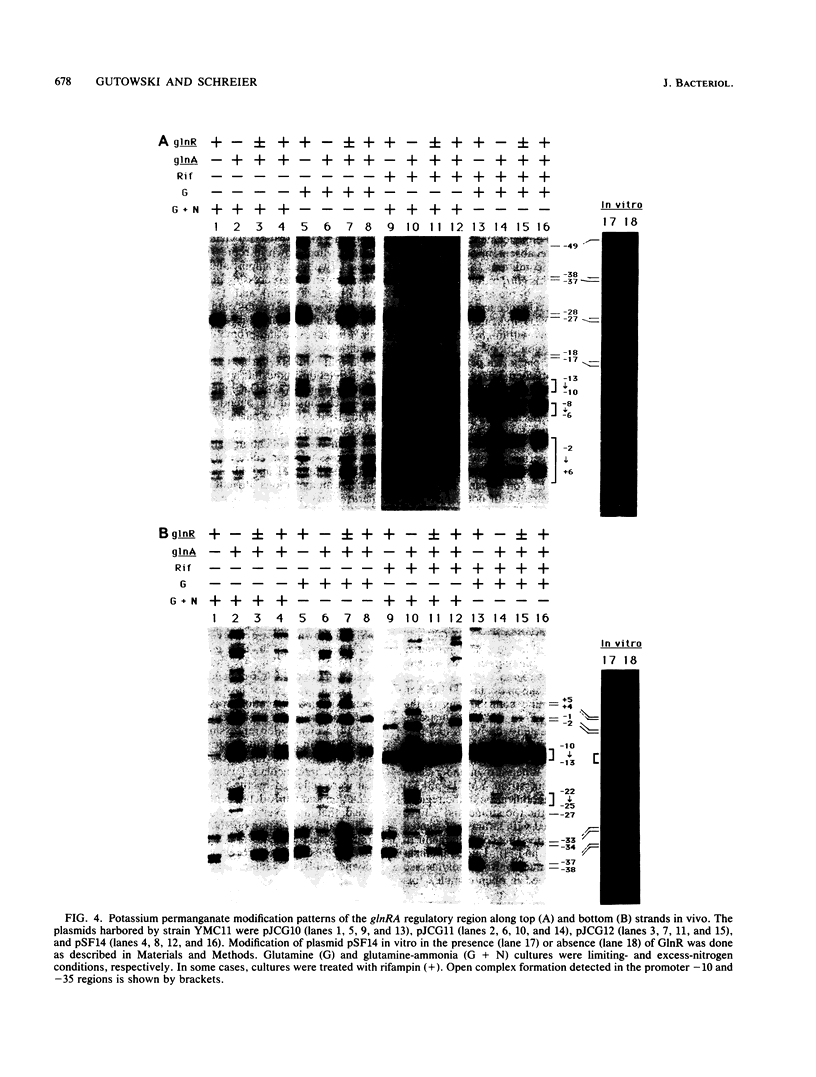
In vivo dimethyl sulfate footprinting of the Bacillus subtilis glnRA regulatory region under repressing and derepressing conditions demonstrated that the GlnR protein, encoded by glnR, interacts with two sites situated within and adjacent to the glnRA promoter. One site, glnRAo1, between positions -40 and -60 relative to the start point of transcription, is a 21-bp symmetrical element that has been identified as essential for glnRA regulation (H. J. Schreier, C. A. Rostkowski, J. F. Nomellini, and K. D. Hirschi, J. Mol. Biol. 220:241-253, 1991). The second site, glnRAo2, is a quasisymmetrical element having partial homology to glnRAo1 and is located within the promoter between positions -17 and -37. The symmetry and extent of modifications observed for each site during repression and derepression indicated that GlnR interacts with the glnRA regulatory region by binding to both sites in approximately the same manner. Experiments using potassium permanganate to probe open complex formation by RNA polymerase demonstrated that transcriptional initiation is inhibited by GlnR. Furthermore, distortion of the DNA helix within glnRAo2 occurred upon GlnR binding. While glutamine synthetase, encoded by glnA, has been implicated in controlling glnRA expression, analyses with dimethyl sulfate and potassium permanganate ruled out a role for glutamine synthetase in directly influencing transcription by binding to operator and promoter regions. Our results suggested that inhibition of transcription from the glnRA promoter involves GlnR occupancy at both glnRAo1 and glnRAo2. In addition, modification of bases within the glnRAo2 operator indicated that control of glnRA expression under nitrogen-limiting (derepressing) conditions included the involvement of a factor(s) other than GlnR.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albertini A. M., Galizzi A. The Bacillus subtilis outB gene is highly homologous to an Escherichia coli ntr-like gene. J Bacteriol. 1990 Sep;172(9):5482–5485. doi: 10.1128/jb.172.9.5482-5485.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backman K., Chen Y. M., Magasanik B. Physical and genetic characterization of the glnA--glnG region of the Escherichia coli chromosome. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3743–3747. doi: 10.1073/pnas.78.6.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowiec J. A., Zhang L., Sasse-Dwight S., Gralla J. D. DNA supercoiling promotes formation of a bent repression loop in lac DNA. J Mol Biol. 1987 Jul 5;196(1):101–111. doi: 10.1016/0022-2836(87)90513-4. [DOI] [PubMed] [Google Scholar]
- Claverie-Martin F., Magasanik B. Role of integration host factor in the regulation of the glnHp2 promoter of Escherichia coli. Proc Natl Acad Sci U S A. 1991 Mar 1;88(5):1631–1635. doi: 10.1073/pnas.88.5.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean D. R., Aronson A. I. Selection of Bacillus subtilis mutants impaired in ammonia assimilation. J Bacteriol. 1980 Feb;141(2):985–988. doi: 10.1128/jb.141.2.985-988.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean D. R., Hoch J. A., Aronson A. I. Alteration of the Bacillus subtilis glutamine synthetase results in overproduction of the enzyme. J Bacteriol. 1977 Sep;131(3):981–987. doi: 10.1128/jb.131.3.981-987.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuel T. F., Ginsburg A., Yeh J., Shelton E., Stadtman E. R. Bacillus subtilis glutamine synthetase. Purification and physical characterization. J Biol Chem. 1970 Oct 25;245(20):5195–5205. [PubMed] [Google Scholar]
- Ferrari F. A., Nguyen A., Lang D., Hoch J. A. Construction and properties of an integrable plasmid for Bacillus subtilis. J Bacteriol. 1983 Jun;154(3):1513–1515. doi: 10.1128/jb.154.3.1513-1515.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher S. H., Rosenkrantz M. S., Sonenshein A. L. Glutamine synthetase gene of Bacillus subtilis. Gene. 1984 Dec;32(3):427–438. doi: 10.1016/0378-1119(84)90018-0. [DOI] [PubMed] [Google Scholar]
- Fisher S. H., Sonenshein A. L. Bacillus subtilis glutamine synthetase mutants pleiotropically altered in glucose catabolite repression. J Bacteriol. 1984 Feb;157(2):612–621. doi: 10.1128/jb.157.2.612-621.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman A. D. Integration of developmental signals and the initiation of sporulation in B. subtilis. Cell. 1991 Apr 5;65(1):5–8. doi: 10.1016/0092-8674(91)90353-z. [DOI] [PubMed] [Google Scholar]
- Haima P., Bron S., Venema G. The effect of restriction on shotgun cloning and plasmid stability in Bacillus subtilis Marburg. Mol Gen Genet. 1987 Sep;209(2):335–342. doi: 10.1007/BF00329663. [DOI] [PubMed] [Google Scholar]
- Hirschman J., Wong P. K., Sei K., Keener J., Kustu S. Products of nitrogen regulatory genes ntrA and ntrC of enteric bacteria activate glnA transcription in vitro: evidence that the ntrA product is a sigma factor. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7525–7529. doi: 10.1073/pnas.82.22.7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Magasanik B. Reversible phosphorylation of an enhancer binding protein regulates the transcription of bacterial nitrogen utilization genes. Trends Biochem Sci. 1988 Dec;13(12):475–479. doi: 10.1016/0968-0004(88)90234-4. [DOI] [PubMed] [Google Scholar]
- Nakano Y., Kato C., Tanaka E., Kimura K., Horikoshi K. Nucleotide sequence of the glutamine synthetase gene (glnA) and its upstream region from Bacillus cereus. J Biochem. 1989 Aug;106(2):209–215. doi: 10.1093/oxfordjournals.jbchem.a122834. [DOI] [PubMed] [Google Scholar]
- Nakano Y., Kimura K. Purification and characterization of a repressor for the Bacillus cereus glnRA operon. J Biochem. 1991 Feb;109(2):223–228. [PubMed] [Google Scholar]
- Ninfa A. J., Reitzer L. J., Magasanik B. Initiation of transcription at the bacterial glnAp2 promoter by purified E. coli components is facilitated by enhancers. Cell. 1987 Sep 25;50(7):1039–1046. doi: 10.1016/0092-8674(87)90170-x. [DOI] [PubMed] [Google Scholar]
- Pan F. L., Coote J. G. Glutamine synthetase and glutamate synthase activities during growth and sporulation in Bacillus subtilis. J Gen Microbiol. 1979 Jun;112(2):373–377. doi: 10.1099/00221287-112-2-373. [DOI] [PubMed] [Google Scholar]
- Popham D. L., Szeto D., Keener J., Kustu S. Function of a bacterial activator protein that binds to transcriptional enhancers. Science. 1989 Feb 3;243(4891):629–635. doi: 10.1126/science.2563595. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasse-Dwight S., Gralla J. D. KMnO4 as a probe for lac promoter DNA melting and mechanism in vivo. J Biol Chem. 1989 May 15;264(14):8074–8081. [PubMed] [Google Scholar]
- Sasse-Dwight S., Gralla J. D. Probing co-operative DNA-binding in vivo. The lac O1:O3 interaction. J Mol Biol. 1988 Jul 5;202(1):107–119. doi: 10.1016/0022-2836(88)90523-2. [DOI] [PubMed] [Google Scholar]
- Sasse-Dwight S., Gralla J. D. Probing the Escherichia coli glnALG upstream activation mechanism in vivo. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8934–8938. doi: 10.1073/pnas.85.23.8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasse-Dwight S., Gralla J. D. Role of eukaryotic-type functional domains found in the prokaryotic enhancer receptor factor sigma 54. Cell. 1990 Sep 7;62(5):945–954. doi: 10.1016/0092-8674(90)90269-k. [DOI] [PubMed] [Google Scholar]
- Schreier H. J., Brown S. W., Hirschi K. D., Nomellini J. F., Sonenshein A. L. Regulation of Bacillus subtilis glutamine synthetase gene expression by the product of the glnR gene. J Mol Biol. 1989 Nov 5;210(1):51–63. doi: 10.1016/0022-2836(89)90290-8. [DOI] [PubMed] [Google Scholar]
- Schreier H. J., Fisher S. H., Sonenshein A. L. Regulation of expression from the glnA promoter of Bacillus subtilis requires the glnA gene product. Proc Natl Acad Sci U S A. 1985 May;82(10):3375–3379. doi: 10.1073/pnas.82.10.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreier H. J., Rostkowski C. A., Nomellini J. F., Hirschi K. D. Identification of DNA sequences involved in regulating Bacillus subtilis glnRA expression by the nitrogen source. J Mol Biol. 1991 Jul 20;220(2):241–253. doi: 10.1016/0022-2836(91)90010-4. [DOI] [PubMed] [Google Scholar]
- Schreier H. J., Sonenshein A. L. Altered regulation of the glnA gene in glutamine synthetase mutants of Bacillus subtilis. J Bacteriol. 1986 Jul;167(1):35–43. doi: 10.1128/jb.167.1.35-43.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenshein A. L., Cami B., Brevet J., Cote R. Isolation and characterization of rifampin-resistant and streptolydigin-resistant mutants of Bacillus subtilis with altered sporulation properties. J Bacteriol. 1974 Oct;120(1):253–265. doi: 10.1128/jb.120.1.253-265.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauch M. A., Aronson A. I., Brown S. W., Schreier H. J., Sonenhein A. L. Sequence of the Bacillus subtilis glutamine synthetase gene region. Gene. 1988 Nov 30;71(2):257–265. doi: 10.1016/0378-1119(88)90042-x. [DOI] [PubMed] [Google Scholar]
- Su W., Porter S., Kustu S., Echols H. DNA-looping and enhancer activity: association between DNA-bound NtrC activator and RNA polymerase at the bacterial glnA promoter. Proc Natl Acad Sci U S A. 1990 Jul;87(14):5504–5508. doi: 10.1073/pnas.87.14.5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedel A., Weiss D. S., Popham D., Dröge P., Kustu S. A bacterial enhancer functions to tether a transcriptional activator near a promoter. Science. 1990 Apr 27;248(4954):486–490. doi: 10.1126/science.1970441. [DOI] [PubMed] [Google Scholar]
- Zhang J., Strauch M., Aronson A. I. Glutamine auxotrophs of Bacillus subtilis that overproduce glutamine synthetase antigen have altered conserved amino acids in or near the active site. J Bacteriol. 1989 Jun;171(6):3572–3574. doi: 10.1128/jb.171.6.3572-3574.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]