Abstract
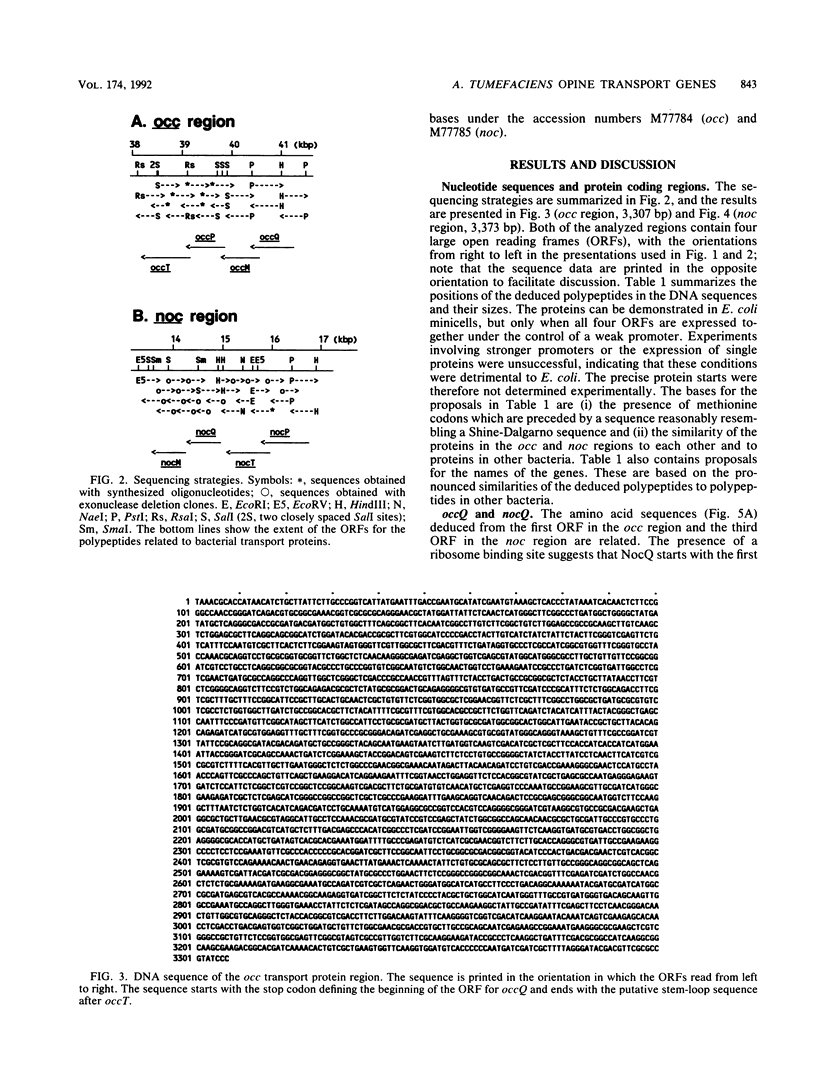
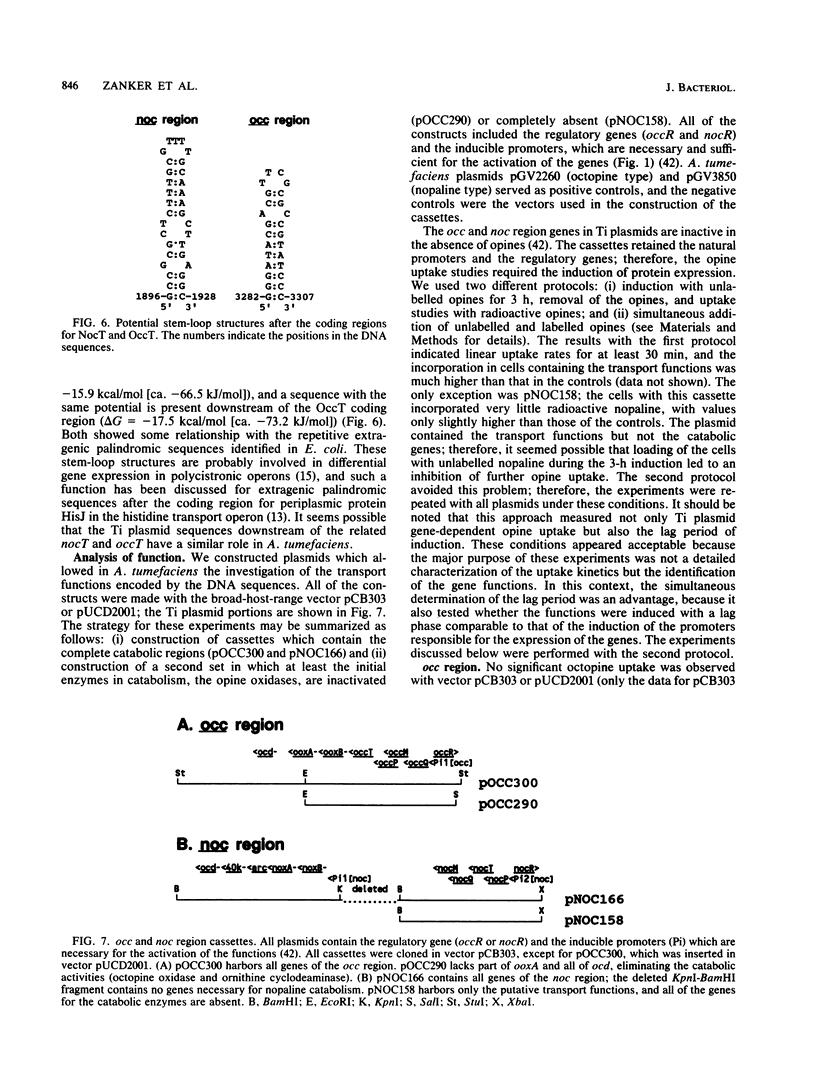
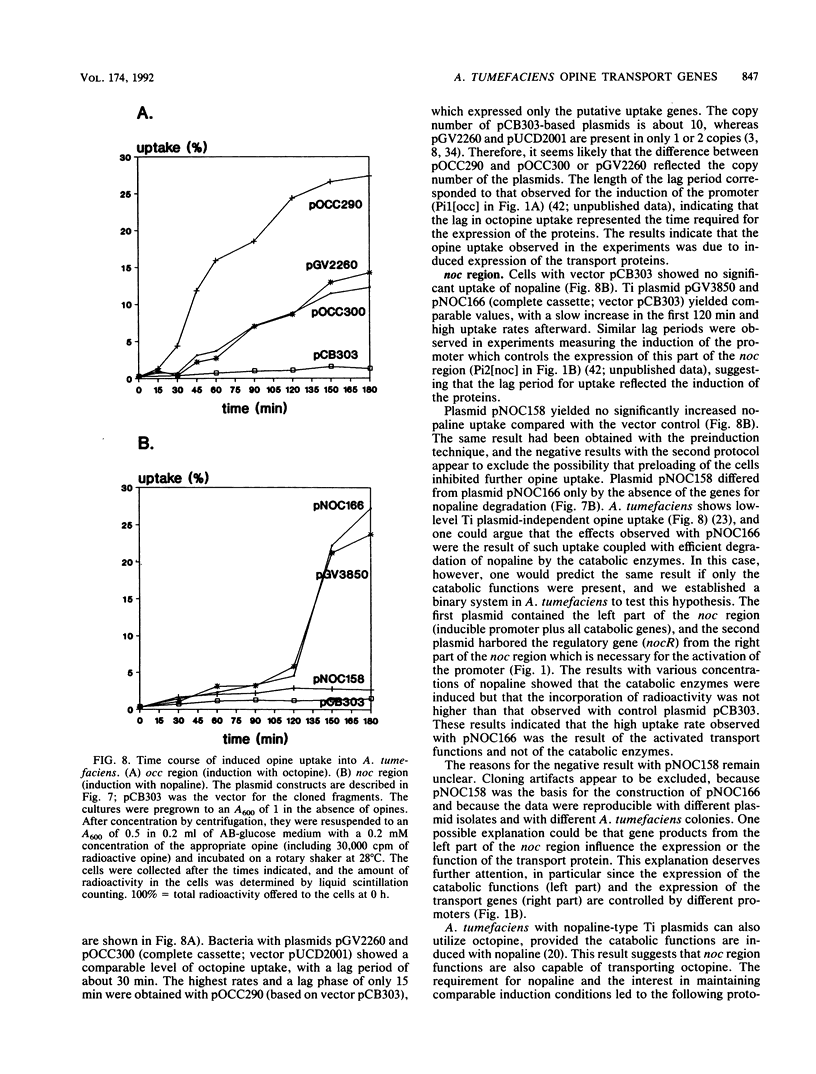
The occ and noc regions of octopine and nopaline Ti plasmids in Agrobacterium tumefaciens are responsible for the catabolic utilization of octopine and nopaline, respectively. Opine-inducible promoters, genes for regulatory proteins and for catabolic enzymes, had been identified in previous work. However, both regions contained additional DNA stretches which were under the control of opine-inducible promoters, but the functions were unknown. We investigated these stretches by DNA sequence and functional analyses. The sequences showed that both of the catabolic regions contain a set of four genes which are transcribed in the same direction. The occ and noc region genes are related, but the arrangement of the genes is different. The deduced polypeptides are related to those of binding protein-dependent transport systems of basic amino acids in other bacteria. The comparison suggested that three of the polypeptides are located in the membrane and that one is a periplasmic protein. We constructed cassettes which contained either the putative transport genes only or the complete occ or noc region; all constructs, however, included the elements necessary for opine-induced expression of the genes (the regulatory gene and the inducible promoters). Uptake studies with 3H-labelled octopine showed that the putative transport genes in the occ region code for octopine uptake proteins. The corresponding studies with 3H-labelled nopaline and the noc region cassettes indicated that the uptake of nopaline requires the putative transport genes and additional functions from the left part of the noc region.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bairoch A. PROSITE: a dictionary of sites and patterns in proteins. Nucleic Acids Res. 1991 Apr 25;19 (Suppl):2241–2245. doi: 10.1093/nar/19.suppl.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron J., Macleod R. A., Dion P. Specificity of octopine uptake by Rhizobium and pseudomonas strains. Appl Environ Microbiol. 1990 May;56(5):1453–1458. doi: 10.1128/aem.56.5.1453-1458.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Greve H., Decraemer H., Seurinck J., Van Montagu M., Schell J. The functional organization of the octopine Agrobacterium tumefaciens plasmid pTiB6s3. Plasmid. 1981 Sep;6(2):235–248. doi: 10.1016/0147-619x(81)90069-x. [DOI] [PubMed] [Google Scholar]
- De Vos G., De Beuckeleer M., Van Montagu M., Schell J. Restriction endonuclease mapping of the octopine tumor-inducing plasmid pTiAch5 of Agrobacterium tumefaciens. Plasmid. 1981 Sep;6(2):249–253. doi: 10.1016/0147-619x(81)90070-6. [DOI] [PubMed] [Google Scholar]
- Deblaere R., Bytebier B., De Greve H., Deboeck F., Schell J., Van Montagu M., Leemans J. Efficient octopine Ti plasmid-derived vectors for Agrobacterium-mediated gene transfer to plants. Nucleic Acids Res. 1985 Jul 11;13(13):4777–4788. doi: 10.1093/nar/13.13.4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessaux Y., Petit A., Tempé J., Demarez M., Legrain C., Wiame J. M. Arginine catabolism in Agrobacterium strains: role of the Ti plasmid. J Bacteriol. 1986 Apr;166(1):44–50. doi: 10.1128/jb.166.1.44-50.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrand S. K., Dessaux Y. Proline biosynthesis encoded by the noc and occ loci of Agrobacterium Ti plasmids. J Bacteriol. 1986 Aug;167(2):732–734. doi: 10.1128/jb.167.2.732-734.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie D. R., Novak S., Kado C. I. Novel high- and low-copy stable cosmids for use in Agrobacterium and Rhizobium. Plasmid. 1985 Sep;14(2):171–175. doi: 10.1016/0147-619x(85)90078-2. [DOI] [PubMed] [Google Scholar]
- Gowrishankar J. Nucleotide sequence of the osmoregulatory proU operon of Escherichia coli. J Bacteriol. 1989 Apr;171(4):1923–1931. doi: 10.1128/jb.171.4.1923-1931.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyon P., Chilton M. D., Petit A., Tempé J. Agropine in "null-type" crown gall tumors: Evidence for generality of the opine concept. Proc Natl Acad Sci U S A. 1980 May;77(5):2693–2697. doi: 10.1073/pnas.77.5.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Higgins C. F., Ames G. F. Two periplasmic transport proteins which interact with a common membrane receptor show extensive homology: complete nucleotide sequences. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6038–6042. doi: 10.1073/pnas.78.10.6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins C. F., Haag P. D., Nikaido K., Ardeshir F., Garcia G., Ames G. F. Complete nucleotide sequence and identification of membrane components of the histidine transport operon of S. typhimurium. Nature. 1982 Aug 19;298(5876):723–727. doi: 10.1038/298723a0. [DOI] [PubMed] [Google Scholar]
- Higgins C. F., Hiles I. D., Whalley K., Jamieson D. J. Nucleotide binding by membrane components of bacterial periplasmic binding protein-dependent transport systems. EMBO J. 1985 Apr;4(4):1033–1039. doi: 10.1002/j.1460-2075.1985.tb03735.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins C. F., McLaren R. S., Newbury S. F. Repetitive extragenic palindromic sequences, mRNA stability and gene expression: evolution by gene conversion? A review. Gene. 1988 Dec 10;72(1-2):3–14. doi: 10.1016/0378-1119(88)90122-9. [DOI] [PubMed] [Google Scholar]
- Hogg R. W. The amino acid sequence of the histidine binding protein of Salmonella typhimurium. J Biol Chem. 1981 Feb 25;256(4):1935–1939. [PubMed] [Google Scholar]
- Holsters M., Silva B., Van Vliet F., Genetello C., De Block M., Dhaese P., Depicker A., Inzé D., Engler G., Villarroel R. The functional organization of the nopaline A. tumefaciens plasmid pTiC58. Plasmid. 1980 Mar;3(2):212–230. doi: 10.1016/0147-619x(80)90110-9. [DOI] [PubMed] [Google Scholar]
- Hynes M. F., Simon R., Pühler A. The development of plasmid-free strains of Agrobacterium tumefaciens by using incompatibility with a Rhizobium meliloti plasmid to eliminate pAtC58. Plasmid. 1985 Mar;13(2):99–105. doi: 10.1016/0147-619x(85)90062-9. [DOI] [PubMed] [Google Scholar]
- Knauf V. C., Nester E. W. Wide host range cloning vectors: a cosmid clone bank of an Agrobacterium Ti plasmid. Plasmid. 1982 Jul;8(1):45–54. doi: 10.1016/0147-619x(82)90040-3. [DOI] [PubMed] [Google Scholar]
- Kraft R., Leinwand L. A. Sequence of the complete P protein gene and part of the M protein gene from the histidine transport operon of Escherichia coli compared to that of Salmonella typhimurium. Nucleic Acids Res. 1987 Oct 26;15(20):8568–8568. doi: 10.1093/nar/15.20.8568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan M., Burgner J. W., Chilton W. S., Gelvin S. B. Transport of nonmetabolizable opines by Agrobacterium tumefaciens. J Bacteriol. 1991 Jan;173(2):903–905. doi: 10.1128/jb.173.2.903-905.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kröger M., Kröger-Block A. Simplified computer programs for search of homology within nucleotide sequences. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):193–201. doi: 10.1093/nar/12.1part1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura C. S., Admon A., Hurt K. A., Ames G. F. The nucleotide-binding site of HisP, a membrane protein of the histidine permease. Identification of amino acid residues photoaffinity labeled by 8-azido-ATP. J Biol Chem. 1990 Nov 15;265(32):19535–19542. [PubMed] [Google Scholar]
- Nohno T., Saito T., Hong J. S. Cloning and complete nucleotide sequence of the Escherichia coli glutamine permease operon (glnHPQ). Mol Gen Genet. 1986 Nov;205(2):260–269. doi: 10.1007/BF00430437. [DOI] [PubMed] [Google Scholar]
- Nonet M. L., Marvel C. C., Tolan D. R. The hisT-purF region of the Escherichia coli K-12 chromosome. Identification of additional genes of the hisT and purF operons. J Biol Chem. 1987 Sep 5;262(25):12209–12217. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sans N., Schindler U., Schröder J. Ornithine cyclodeaminase from Ti plasmid C58: DNA sequence, enzyme properties and regulation of activity by arginine. Eur J Biochem. 1988 Apr 5;173(1):123–130. doi: 10.1111/j.1432-1033.1988.tb13975.x. [DOI] [PubMed] [Google Scholar]
- Sans N., Schröder G., Schröder J. The Noc region of Ti plasmid C58 codes for arginase and ornithine cyclodeaminase. Eur J Biochem. 1987 Aug 17;167(1):81–87. doi: 10.1111/j.1432-1033.1987.tb13306.x. [DOI] [PubMed] [Google Scholar]
- Schardl C. L., Kado C. I. A functional map of the nopaline catabolism genes on the Ti plasmid of Agrobacterium tumefaciens C58. Mol Gen Genet. 1983;191(1):10–16. doi: 10.1007/BF00330882. [DOI] [PubMed] [Google Scholar]
- Schindler U., Sans N., Schröder J. Ornithine cyclodeaminase from octopine Ti plasmid Ach5: identification, DNA sequence, enzyme properties, and comparison with gene and enzyme from nopaline Ti plasmid C58. J Bacteriol. 1989 Feb;171(2):847–854. doi: 10.1128/jb.171.2.847-854.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider K., Beck C. F. New expression vectors for identifying and testing signal structures for initiation and termination of transcription. Methods Enzymol. 1987;153:452–461. doi: 10.1016/0076-6879(87)53071-3. [DOI] [PubMed] [Google Scholar]
- Schrell A., Alt-Moerbe J., Lanz T., Schroeder J. Arginase of Agrobacterium Ti plasmid C58. DNA sequence, properties, and comparison with eucaryotic enzymes. Eur J Biochem. 1989 Oct 1;184(3):635–641. doi: 10.1111/j.1432-1033.1989.tb15060.x. [DOI] [PubMed] [Google Scholar]
- Stachel S. E., An G., Flores C., Nester E. W. A Tn3 lacZ transposon for the random generation of beta-galactosidase gene fusions: application to the analysis of gene expression in Agrobacterium. EMBO J. 1985 Apr;4(4):891–898. doi: 10.1002/j.1460-2075.1985.tb03715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zagursky R. J., Berman M. L. Cloning vectors that yield high levels of single-stranded DNA for rapid DNA sequencing. Gene. 1984 Feb;27(2):183–191. doi: 10.1016/0378-1119(84)90139-2. [DOI] [PubMed] [Google Scholar]
- Zambryski P., Joos H., Genetello C., Leemans J., Montagu M. V., Schell J. Ti plasmid vector for the introduction of DNA into plant cells without alteration of their normal regeneration capacity. EMBO J. 1983;2(12):2143–2150. doi: 10.1002/j.1460-2075.1983.tb01715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Lintig J., Zanker H., Schröder J. Positive regulators of opine-inducible promoters in the nopaline and octopine catabolism regions of Ti plasmids. Mol Plant Microbe Interact. 1991 Jul-Aug;4(4):370–378. doi: 10.1094/mpmi-4-370. [DOI] [PubMed] [Google Scholar]



