Abstract
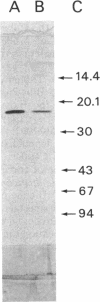
The final enzymatic reaction in the conversion of precorrin-6x to hydrogenobyrinic acid by cell-free protein preparations from Pseudomonas denitrificans was shown to be inhibited by hydrogenobyrinic acid. Use was made of this property to prepare the last biosynthetic precursor of hydrogenobyrinic acid, named precorrin-8x. Double-labeling experiments, mass spectrometry, and UV-visible light spectroscopy studies established that precorrin-8x was at the oxidation level of a corrin and differed from precorrin-6x by two additional methyl groups (presumably at C-5 and C-15) and decarboxylation of the acetic acid side chain at C-12. Precorrin-8x was not a corrin but had the same mass as hydrogenobyrinic acid, thus showing that this latter compound is synthesized from the former by a rearrangement. The enzyme catalyzing this rearrangement was purified 80-fold to homogeneity from a recombinant strain of P. denitrificans, sequenced at its N terminus, and shown to be encoded by the cobH gene. It was identical to the previously described hydrogenobyrinic acid-binding protein (F. Blanche, D. Thibaut, D. Frechet, M. Vuilhorgne, J. Crouzet, B. Cameron, G. Müller, K. Hlineny, U. Traub-Eberhard, and M. Zboron, Angew. Chem. Int. Ed. Engl. 29:884-886, 1990). This enzyme had a Km of 0.91 +/- 0.04 microM and a Vmax of 230 nmol h-1 mg-1 at pH 7.7 and was competitively inhibited by hydrogenobyrinic acid with a Ki of 0.17 +/- 0.01 microM. It is proposed that the cobH gene product is a mutase which transfers the methyl group from C-11 to C-12.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bagdasarian M., Lurz R., Rückert B., Franklin F. C., Bagdasarian M. M., Frey J., Timmis K. N. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene. 1981 Dec;16(1-3):237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- Berhauer K., Wagner F., Michna H., Rapp P., Vogelmann H. Zur Chemie und Biochemie der Corrinoide, XXIX. Biogenesewege von der Cobyrinsäure zur Cobysäure und zum Cobinamid bei Propionibacterium shermanii. Hoppe Seylers Z Physiol Chem. 1968 Oct;349(10):1297–1309. [PubMed] [Google Scholar]
- Blanche F., Debussche L., Thibaut D., Crouzet J., Cameron B. Purification and characterization of S-adenosyl-L-methionine: uroporphyrinogen III methyltransferase from Pseudomonas denitrificans. J Bacteriol. 1989 Aug;171(8):4222–4231. doi: 10.1128/jb.171.8.4222-4231.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanche F., Famechon A., Thibaut D., Debussche L., Cameron B., Crouzet J. Biosynthesis of vitamin B12 in Pseudomonas denitrificans: the biosynthetic sequence from precorrin-6y to precorrin-8x is catalyzed by the cobL gene product. J Bacteriol. 1992 Feb;174(3):1050–1052. doi: 10.1128/jb.174.3.1050-1052.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanche F., Thibaut D., Couder M., Muller J. C. Identification and quantitation of corrinoid precursors of cobalamin from Pseudomonas denitrificans by high-performance liquid chromatography. Anal Biochem. 1990 Aug 15;189(1):24–29. doi: 10.1016/0003-2697(90)90038-b. [DOI] [PubMed] [Google Scholar]
- Blanche F., Thibaut D., Famechon A., Debussche L., Cameron B., Crouzet J. Precorrin-6x reductase from Pseudomonas denitrificans: purification and characterization of the enzyme and identification of the structural gene. J Bacteriol. 1992 Feb;174(3):1036–1042. doi: 10.1128/jb.174.3.1036-1042.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron B., Briggs K., Pridmore S., Brefort G., Crouzet J. Cloning and analysis of genes involved in coenzyme B12 biosynthesis in Pseudomonas denitrificans. J Bacteriol. 1989 Jan;171(1):547–557. doi: 10.1128/jb.171.1.547-557.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Martinez-Arias A., Shapira S. K., Chou J. Beta-galactosidase gene fusions for analyzing gene expression in escherichia coli and yeast. Methods Enzymol. 1983;100:293–308. doi: 10.1016/0076-6879(83)00063-4. [DOI] [PubMed] [Google Scholar]
- Crouzet J., Cameron B., Cauchois L., Rigault S., Rouyez M. C., Blanche F., Thibaut D., Debussche L. Genetic and sequence analysis of an 8.7-kilobase Pseudomonas denitrificans fragment carrying eight genes involved in transformation of precorrin-2 to cobyrinic acid. J Bacteriol. 1990 Oct;172(10):5980–5990. doi: 10.1128/jb.172.10.5980-5990.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouzet J., Levy-Schil S., Cameron B., Cauchois L., Rigault S., Rouyez M. C., Blanche F., Debussche L., Thibaut D. Nucleotide sequence and genetic analysis of a 13.1-kilobase-pair Pseudomonas denitrificans DNA fragment containing five cob genes and identification of structural genes encoding Cob(I)alamin adenosyltransferase, cobyric acid synthase, and bifunctional cobinamide kinase-cobinamide phosphate guanylyltransferase. J Bacteriol. 1991 Oct;173(19):6074–6087. doi: 10.1128/jb.173.19.6074-6087.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debussche L., Thibaut D., Cameron B., Crouzet J., Blanche F. Purification and characterization of cobyrinic acid a,c-diamide synthase from Pseudomonas denitrificans. J Bacteriol. 1990 Nov;172(11):6239–6244. doi: 10.1128/jb.172.11.6239-6244.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann H. C., Cagen L. M. Microbial biosynthesis of B12-like compounds. Annu Rev Microbiol. 1970;24:159–208. doi: 10.1146/annurev.mi.24.100170.001111. [DOI] [PubMed] [Google Scholar]
- Leong S. A., Ditta G. S., Helinski D. R. Heme biosynthesis in Rhizobium. Identification of a cloned gene coding for delta-aminolevulinic acid synthetase from Rhizobium meliloti. J Biol Chem. 1982 Aug 10;257(15):8724–8730. [PubMed] [Google Scholar]
- Thibaut D., Blanche F., Debussche L., Leeper F. J., Battersby A. R. Biosynthesis of vitamin B12: structure of precorrin-6x octamethyl ester. Proc Natl Acad Sci U S A. 1990 Nov;87(22):8800–8804. doi: 10.1073/pnas.87.22.8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibaut D., Couder M., Crouzet J., Debussche L., Cameron B., Blanche F. Assay and purification of S-adenosyl-L-methionine:precorrin-2 methyltransferase from Pseudomonas denitrificans. J Bacteriol. 1990 Nov;172(11):6245–6251. doi: 10.1128/jb.172.11.6245-6251.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibaut D., Debussche L., Blanche F. Biosynthesis of vitamin B12: isolation of precorrin-6x, a metal-free precursor of the corrin macrocycle retaining five S-adenosylmethionine-derived peripheral methyl groups. Proc Natl Acad Sci U S A. 1990 Nov;87(22):8795–8799. doi: 10.1073/pnas.87.22.8795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]