Abstract
CD4+ T helper cells differentiate into Th1 or Th2 effector lineages, which orchestrate immunity to different types of microbes. Both Th1 and Th2 differentiation can be induced by Notch, but what dictates which of these programs is activated in response to Notch is not known. The requirement for Notch in T helper differentiation is controversial. Using T cell specific gene ablation of the Notch effector RBP-J or the Notch1 and 2 receptors, we show here that Notch is required on CD4+ T cells for physiological Th2 responses to parasite antigens. We find that Gata3 is necessary for Notch induced Th2 differentiation and identify an upstream gata3 promoter as a direct target for Notch signaling. Moreover, we observed that absence of Gata3 turns Notch from a Th2 inducer into a powerful inducer of Th1 differentiation. Therefore, Gata3 is a critical element determining inductive Th2 differentiation and limiting Th1 differentiation by Notch.
Introduction
Effective immunity against different classes of microorganisms is directed by specialized effector CD4+ T helper subsets, which produce distinct sets of cytokines to activate the effector mechanisms appropriate to combat the microorganisms encountered. The most extensively studied of these T helper subsets are called Th1 and Th2 (Mosmann and Coffman, 1989). Th1 cells are characterized by production of IFNγ and responsible for generating immunity against intracellular pathogens. On the other hand, Th2 cells produce IL4, IL5 and IL13, are necessary for protection against helminth worms, and are involved in allergic reactions (Abbas et al., 1996). For example, exposure to eggs of the parasitic helminth Schistosoma mansoni elicits a particularly strong Th2 response, which is necessary for host survival (Fallon et al., 2000; Pearce et al., 2004). Recently a third effector subset of T helper cells has been identified, characterized by the production of IL17, but much less is known about these cells (reviewed in Weaver et al., 2007).
The different types of effector T helper cells derive from a common precursor, the mature naïve CD4 T cell. Skewing of CD4+ T cells into different lineages depends on signals provided by antigen presenting cells (APC) (Moser and Murphy, 2000). Upon recognition of hallmark features of microorganisms APC produce distinct differentiation signals (Kapsenberg, 2003), which instruct naïve T cells to differentiate into the lineage best suited to fight the particular class of microbial threat encountered. A well characterized APC-derived differentiation signal is IL12, which promotes differentiation of naïve CD4 T cells into the Th1 lineage (Kapsenberg, 2003; Moser and Murphy, 2000). Several APC derived Th2 promoting signals have been described, which include OX40 ligand, IL6 and the Notch ligand Jagged (Amsen et al., 2004; Flynn et al., 1998; Ito et al., 2005; Mowen and Glimcher, 2004; Rincon et al., 1997; So et al., 2006).
Two transcription factors regulate differentiation into the Th1 and Th2 lineages, called Tbet and Gata3, respectively (Lee et al., 2000; Mullen et al., 2001; Ouyang et al., 1998; Szabo et al., 2000; Zhang et al., 1997; Zheng and Flavell, 1997). These factors not only promote differentiation into their respective lineages, but also limit differentiation into the other lineage by antagonizing the opposing transcription factor (Hwang et al., 2005; Usui et al., 2003; Usui et al., 2006). Ectopic expression of Gata3 is sufficient for activation of that program (Lee et al., 2000; Murphy and Reiner, 2002; Ouyang et al., 2000), while deletion of the gata3 gene blocks differentiation into the Th2 lineage (Pai et al., 2004; Yamashita et al., 2004; Zhu et al., 2004). Little is known about the signals regulating expression of the gata3 gene. Expression of Gata3 can be induced by IL4 receptor signaling, in a STAT6 dependent manner, consistent with the ability of IL4 to induce Th2 differentiation (Kurata et al., 1999; Murphy and Reiner, 2002; Ouyang et al., 2000). How this pathway connects with the gata3 gene remains to be determined. Importantly, other signals than IL4 must be able to drive Th2 differentiation and thus expression of gata3. The initial source of IL4 for Th2 differentiation in vivo has not been identified (Ansel et al., 2006) and Th2 responses can be generated when only T cells can make IL4, arguing against a requisite role for an external source of IL4 in Th2 responses (Schmitz et al., 1994). Finally, Th2 responses do occur, albeit with reduced vigor, under conditions where IL4 receptor signaling is prevented (Ansel et al., 2006; Finkelman et al., 2000; Jankovic et al., 2000; Noben-Trauth et al., 1997). Correspondingly, although maximal expression of Gata3, at least in vitro, does require STAT6, Gata3 is expressed in STAT6 deficient T cells at higher levels than in Th1 cells (Ouyang et al., 2000). While this expression involves on an auto activation loop, in which Gata3 maintains its own expression, the pathways responsible for the initial STAT6 independent expression have not been identified (Ouyang et al., 2000).
Recently the Notch family has been implicated in T helper differentiation (Maillard et al., 2005). Notch is a heterodimeric surface receptor consisting of an extracellular ligand binding chain noncovalently associated with a transmembrane polypeptide with a long intracellular tail (Bray, 2006). Mammals have 4 different Notch genes, of which Notch1 and 2 are the most similar to one another, while Notch3 and 4 are more divergent (Maillard et al., 2005). Two conserved families of ligands for Notch exist, called Jagged and Delta, encoded by two and three separate genes, respectively, in mammals (Maillard et al., 2005). These families are structurally quite different and have both overlapping and distinct functions (de La Coste and Freitas, 2006). Upon binding ligand Notch undergoes sequential proteolytic cleavages, including one catalyzed by a gamma secretase complex, which result in the release of the intracellular domain (NICD) from the membrane (Artavanis-Tsakonas et al., 1999). In the canonical signaling pathway used by all 4 Notchs, this NICD translocates to the nucleus where it associates with the DNA binding factor RBP-J (also known as CBF1 or CSL) believed to be pre-bound to its target site on DNA (Artavanis-Tsakonas et al., 1999). Binding of the NICD to RBP-J results in displacement of transcriptional corepressors from RBP-J (Maillard et al., 2005) and recruitment of the Mastermind protein. The resulting trimolecular complex in turn recruits various transcriptional co-activators (Maillard et al., 2005), thus converting RBP-J from a transcriptional repressor into a transactivator.
Notch is known for its role in binary cell fate decisions in many different developmental processes (Bray, 2006). In some settings, Notch regulates such decisions by a lateral inhibition mechanism, in which adoption of a primary fate is inhibited by Notch signaling, allowing cells to differentiate into a secondary fate by default. In other settings Notch acts through inductive signaling by actively promoting expression of lineage differentiation genes (Bray, 2006).
No consensus exists about the role of Notch in T helper differentiation. In gain of function experiments Notch promoted either Th1 or Th2 differentiation, (Amsen et al., 2004; Maekawa et al., 2003; Minter et al., 2005), but only Th2 responses were blocked in RBP-J deficient mice or mice expressing a dominant negative Mastermind transgene (Amsen et al., 2004; Tanigaki et al., 2004; Tu et al., 2005). In sharp contrast, inhibition of activation of Notch itself, with chemical inhibitors of the gamma secretase or a soluble Delta-Fc fusion protein, resulted in inhibition of Th1 but not Th2 responses (Minter et al., 2005). One explanation for these apparent discrepancies could be that Th2 responses depend on a Notch independent function of RBP-J, for which evidence exists (Barolo et al., 2000). In general, all the approaches used in this field (including ours) have relied on inhibition of Notch activity by indirect methods that may affect signaling pathways not depending on Notch. Gamma secretase cleaves membrane molecules other than Notch (Wolfe and Kopan, 2004) and prominent Notch independent functions exist for Mastermind (Katada et al., 2006; Shen et al., 2006) and RBP-J (Barolo et al., 2000). To resolve the controversy it is necessary, therefore, to use direct loss of function approaches of the essential components of the pathway, RBPJ and the Notch genes themselves, and to establish the mechanism whereby Notch mediates its effects.
In the present study we examined T helper 2 differentiation using mice carrying T cell specific deletions of RBP-J or the notch1 and notch2 genes themselves. We established that Notch is required for Th2 responses to parasite antigens under physiological conditions and reveal that Notch drives Th2 differentiation by an inductive mechanism through direct trans activation of the gata3 gene. Furthermore, we report that Gata3 acts as a switch factor, as its absence converts Notch from a Th2 inducer into a powerful inducer of Th1 differentiation.
Results
RBP-J is required for Th2 responses in vivo
Some studies found the Notch pathway required for Th2 responses, while others did not. A Th2 defect was previously found in mice with a T cell specific deletion of the rbpj gene under weak Th2 inducing conditions (Tanigaki et al., 2004). It has not been tested whether this factor is required under strong and physiologically important Th2 inducing conditions, such as those elicited by parasite antigens.
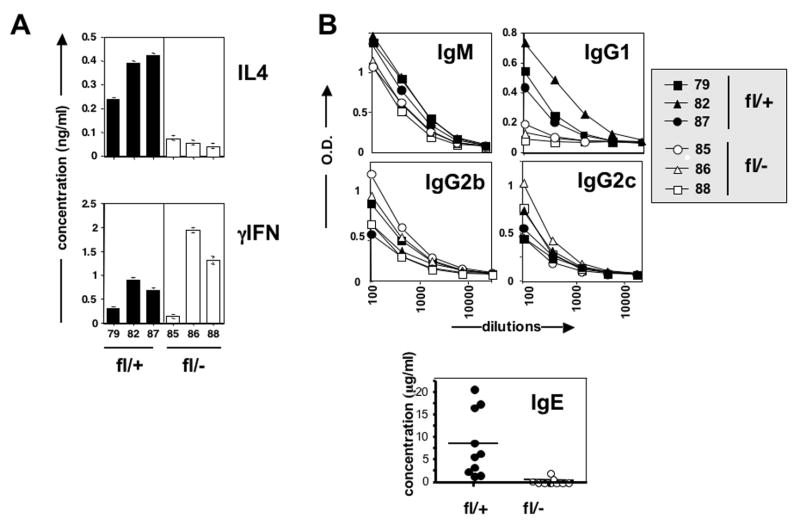
Therefore, we immunized rbpj deficient mice with extract of the eggs obtained from Schistosoma mansoni, which induce pronounced Th2 type immunity during normal infection (Pearce et al., 2004). Extracts from such eggs contain potent adjuvant activity and have been used extensively to study Th2 responses in vivo (Pearce et al., 2004). Indeed, immunization with these extracts elicited strong Th2 responses as witnessed by the production of Th2 dependent antibody isotypes IgG1 and IgE, as well as by the presence in spleens of these mice of antigen specific CD4 T cells secreting the signature Th2 cytokine IL4 upon in vitro rechallenge (Figure 1). Strikingly, both Th2 dependent antibody production and secretion of IL4 were abrogated in mice lacking RBP-j expression in T cells (Figure 1), documenting the in vivo requirement for the Notch pathway in Th2 responses. In contrast, production of IgM, which is partially dependent on CD4+ T cell help, but not on a particular effector type, was not significantly affected by rbpjk deficiency. Although SEA does not elicit strong Th1 responses, some Th1 dependent antibodies (IgG2b and IgG2c) and cytokine (IFNγ) could be measured in these immunized mice (Figure 1). We did not, however, find a consistent effect from RBP-J deficiency on this type of response. Thus, although these data do not exclude a role for the Notch pathway in Th1 induction (see discussion), they do reinforce the notion that this pathway is required for Th2 responses.
Figure 1. Generation of Th2 mediated immunity in vivo is dependent on RBP-J.
RBP-J deficient (RBPflox/−/CD4Cre) (white bars and symbols) or heterozygous (RBPflox/+/CD4Cre) littermates (black bars and symbols) were immunized with extract from Schistosoma mansoni eggs (SEA). Cytokine responses of purified splenic CD4+ T cells were measured upon in vitro restimulation with SEA (a). Each bar represents the response of CD4+ T cells from an individual mouse. In addition, sera from individual heterozygous control (closed symbols) or RBP-J deficient mice (open symbols) were tested for the presence of various isotypes of antibodies against SEA (b). Data in (a) and (b) are representative of 5 separate experiments. For IgE, values obtained from several experiments are shown combined, with each dot representing a single mouse.
Th2 responses depend on expression of Notch 1 and 2
It is conceivable that the results presented above are a reflection of a Notch independent function of RBP-J (Barolo et al., 2000). Consistent with this idea, inhibition of activation of Notch itself using gamma secretase inhibitors did not perturb Th2 responses in vitro (Minter et al., 2005). We therefore determined whether deficiency for Notch affects Th2 differentiation.
Of the 4 mammalian Notch’s, Notch 1 and 2 are phylogenetically closest to one another, while Notch3 and 4 are more divergent (Maillard et al., 2005). Mice lacking individual Notch genes have no defect in T helper differentiation (Tacchini-Cottier et al., 2004) (and our unpublished observations). We therefore examined whether double deficiency in Notch1 and 2, the Notch genes expressed in naïve CD4+ T cells (Amsen et al., 2004), affects T helper responses in the presence of SEA as a strong Th2 adjuvant. Deletion of Notch1 and 2 by CD4 promoter driven Cre does not affect thymic development (unpublished results), allowing the study of peripheral T helper responses. To this end, we cultured AND TCR transgenic naïve CD4+ T cells, derived from mice carrying T cell specific deletions of both notch1 and 2 genes, in vitro with SEA treated APC and pigeon cytochrome C. After 5 days we assayed T helper differentiation by intracellular cytokine staining for IL4 and IFNγ. Strikingly, Notch1/2 deficient CD4+ T cells failed to make IL4, while no consistent effect was found on production of IFNγ (Figure 2a). Thus, Notch1 and 2 are essential for induction of Th2 differentiation under strong Th2 inducing conditions, such as those created by SEA treated APC. In contrast, no effect from Notch1/2 deficiency was found in standard in vitro Th1 and Th2 differentiation paradigms, in which differentiation is induced by the addition of cytokines (IL12 or IL4) and neutralizing antibodies (anti IL4 or anti IFNγ) (Figure 2b). Collectively, these data reveal that the Notch pathway is required for induction of Th2 responses under physiological conditions. This role is obscured, however, in standard in vitro T helper cultures, where exogenous cytokines likely override the physiological mechanisms, presumably explaining the previously reported inability of gamma secretase inhibitors to block Th2 differentiation (Minter et al., 2005).
Figure 2. Notch1 and 2 are required for physiological Th2 responses.
(a) Naïve CD4+ T cells were isolated from AND TCR transgenic Notch1/2 double deficient mice (Notch1flox/flox/Notch2flox/flox CD4Cre) (open dots) or wild type (Notch1flox/+/Notch2flox/+ CD4Cre) (closed dots) littermates and stimulated in vitro with collagenase treated splenic APC in the presence of SEA (20μg/ml) and pigeon cytochrome C (10μg/ml). After 5 days viable effector cells were restimulated with PMA and ionomycin and IL4 and IFNγ production was measured by intracellular cytokine staining. Each dot represents percentage IL4+ or IFNγ+ cells in cells obtained from an individual mouse. Data represent cumulative results from 3 independent experiments.
(b) Naïve CD4+ T cells were isolated from Notch1/2 double deficient (hatched bars) or wild type (filled bars) littermates (all positive for the CD4 Cre transgene) and activated in vitro with soluble anti CD3 and anti CD28 and splenic APC in the presence of Th1 (IL12 and anti IL4) or Th2 (IL4 and anti IFNγ) polarizing conditions. After 5 days viable effector cells were harvested, restimulated with plate bound anti CD3 (10μg/ml) and 48 hour supernatants were assayed for IL4 and IFNγ. Cytokine concentrations from cells obtained from individual mice are shown.
Promoter specific regulation of the gata3 gene by Notch
In mice lacking RBP-J decreased Th2 responses were accompanied by increased Th1 responses (Tanigaki et al., 2004). Thus, Notch might promote Th2 mediated immunity as a default consequence of interfering with Th1 induction (Tu et al., 2005). Alternatively, Notch may actively promote Th2 differentiation in an inductive manner, in which case the increased Th1 responses in RBP-J deficient mice (Tanigaki et al., 2004) result from a lack of cross inhibition by Th2 factors.
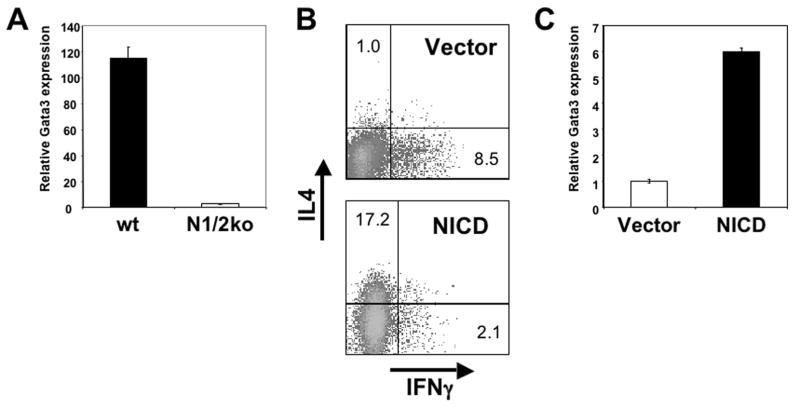
The most conclusive evidence for an inductive mechanism would be to establish that key Th2 differentiation genes are regulated directly by Notch. The central factor for the differentiation of Th2 cells is Gata3 (Lee et al., 2000; Ouyang et al., 2000; Ouyang et al., 1998; Pai et al., 2004; Yamashita et al., 2004; Zhang et al., 1997; Zheng and Flavell, 1997; Zhu et al., 2004). As shown in Figure 3a, under physiological conditions expression of this factor is dependent on Notch, as it is abrogated in AND TCR transgenic Notch1/2 double deficient T cells activated with antigen in the presence of SEA (Figure 3a). To examine whether Notch regulates expression of the gata3 gene in a direct fashion, we introduced an activated Notch allele (NICD) in CD4 T cells. Expression of this allele, consisting of the intracellular domain of Notch, results in Th2 differentiation (Amsen et al., 2004). While this result was not obtained in another study (Maekawa et al., 2003), there this effect was likely obscured due to the use of a mixed T cell population (including memory/effector cells) from naturally Th2 prone Balb/c T cells. In fact, NICD induces Th2 differentiation even in STAT6 deficient T cells (Figure 3b). NICD also induced expression of the gata3 gene in STAT6 deficient T cells (Figure 3c). As STAT6 deficiency excludes auto/paracrine effects from elevated IL4 production induced by NICD, these results are consistent with a direct link between Notch and Gata3.
Figure 3. (a) Notch is required for Gata3 expression.
Naïve CD4+ T cells from wild type (closed bar) or Notch1/2 deficient mice (open bar) were isolated and stimulated as in Figure 2(a). After 5 days in culture, cDNA was prepared and tested for the abundance of gata3 transcripts.
(b) Notch ICD induces IL4 independent Th2 differentiation. Naïve STAT6 −/− CD4+ T cells were activated with splenic APC and soluble antibodies to CD3 and CD28 and transduced with a retrovirus encoding NICD linked to GFP through an IRES sequence (right panel) or with control virus (left panel). 3 days after transduction viable effector cells were restimulated with PMA and ionomycin and IL4 and IFNγ production was measured by intracellular cytokine staining. Numbers represent percentages in each quadrant.
(c) Notch ICD induces IL4 independent Gata3 expression. Naïve STAT6 −/− CD4+ T cells were activated and transduced as in (b). 3 days after transduction GFP+ cells were isolated by FACSorting, cDNA was prepared and the abundance of gata3 transcripts was determined by real time PCR. Open bar represents Vector transduced and closed bar represents NICD transduced cells.
We then studied whether Notch regulates global activity of this gene, or the activity of a specific promoter. Expression of the gata3 gene is controlled by two different promoters, separated by approximately 10 kB (Asnagli et al., 2002). Each of these drives expression of a transcript containing a unique first exon, 1a or 1b respectively, which splices to a common exon 2. Both these exons 1 contain only 5′UTR sequence; the translational start site is present in the common second exon. Thus, both transcripts encode for the same Gata3 protein. The usage of distinct first exons by the different promoters allows specific measurement of the activities of these promoters separately in their native chromatin contexts. We therefore measured the induction of both transcripts by NICD, again using STAT6 deficient CD4+ T cells. Strikingly, while Notch did not significantly affect expression of exon1b, expression of exon 1a was strongly induced (Figure 4a). Notch responsiveness of exon 1a was abrogated in RBP-J deficient CD4+ T cells (Figure 4b), consistent with the requirement for this Notch effector in Th2 differentiation (Figure 1). Thus, Notch does not affect global activity of the gata3 gene, but specifically activates the upstream gata3 promoter in an RBP-J dependent manner.
Figure 4. (a) Notch stimulates transcriptional activity of the upstream but not the downstream Gata3 promoter.
Naïve STAT6 −/− CD4+ T cells were transduced, sorted for GFP+ cells and cDNA was prepared as in 3(c). The abundance of exon1a or exon 1b containing transcripts was determined by real time PCR using a common 3′ primer annealing in exon 2 and specific primers annealing in exon 1a (left) or exon 1b (right).(b) Notch induced activity of the upstream Gata3 promoter is dependent on RBP-J. Naïve CD4+ T cells from wild type or RBP-J deficient mice were transduced and analyzed as in (a) for the abundance of exon 1a (top) or exon 1b (bottom) containing transcripts. Neutralizing antibody to IL4 (10μg/ml) was included in the cultures.
A physical link between the Notch pathway and the gata3 gene
To determine whether the upstream gata3 promoter is a direct target of Notch we scanned it for RBP-J binding sites. A potential RBP-J binding element was found (Figure 5a). This site is conserved between mice and humans, suggesting it may be an important site. No RBP-J binding sequence was found in the downstream promoter. The putative RBP-J element in the upstream gata3 promoter (hereafter referred to as RG3P1a) corresponds to one published RBP-J consensus binding sequence (Bailey and Posakony, 1995), but not to another (Tun et al., 1994). To determine whether RG3P1a is a bona fide RBP-J binding element we used double strand DNA oligonucleotides containing RG3P1a in EMSA with extracts from cells transfected with RBP-J. Indeed, a shifted complex was obtained when radiolabeled RG3P1a was incubated with extracts from cells transfected with RBP-J, but not with extracts from cells transfected with control vector (Figure 5b). No complex was formed either with extracts from cells transfected with RBP-J R218H, which contains a single amino acid substitution perturbing its ability to bind DNA (Kato et al., 1997). Furthermore, no complex was formed upon incubation of RBP-J containing lysate with radiolabeled oligos containing a point mutation which precludes binding of RBP-J (mRG3P1a) (Figure 5b). Formation of the radioactive RBP-J-RG3P1a complex could be inhibited by addition of excess unlabeled RG3P1a, or oligos encompassing a previously characterized RBP-J binding element (RE) (Tun et al., 1994), but not by addition of mRG3P1a. Finally, binding activity to RG3P1a was found in extracts from CD4+ T cells and identified as RBP-J by supershift using an antibody to RBP-J (Figure 5c). These data establish RG3P1a as a bona fide RBP-J binding element.
Figure 5. The upstream Gata3 promoter contains a bona fide conserved RBP-J binding element.
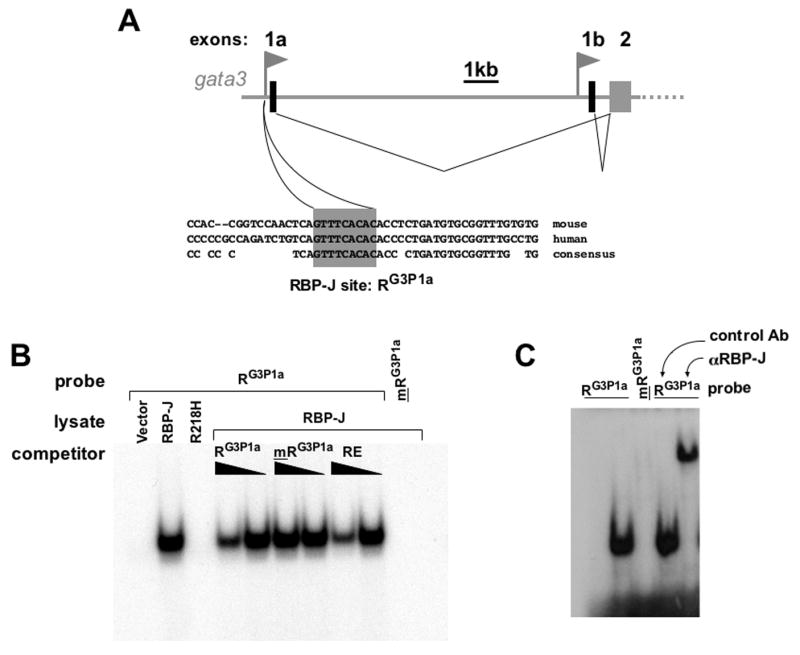
(a) diagram of the Gata3 locus showing upstream and downstream Gata3 promoters each driving expression of a unique exon 1, which is spliced to the common exon 2. Underneath, the consensus RBP-J binding element is shown and its conservation between mouse and human. (b) EMSA using radiolabeled double strands probe containing the RBPG3P1a and lysates from CHO cells transfected with vector alone (1st lane), RBP-J (2nd lane and 4th to 10th lane) or RBP-J R218H (3rd lane). A probe carrying a point mutation mRBPG3P1a was used as specificity control (10th lane). The specific RBP-J-RBPG3P1a complex was competed away by addition of (10-fold or 3-fold) excess unlabeled RBPG3P1a (lanes 4 and 5) or the previously described RBP-J binding element RE (lanes 8 and 9), but not by the mutant probe mRBPG3P1a (lanes 6 and 7). (c) EMSA using lysates from CD4+ T cells (lanes 2 to 5) and radiolabeled RBPG3P1a, (lanes 1,2 and 4,5) or mRBPG3P1a(lane 3). Antibody to RBP-J (lane 5) or control mouse Ig (lane 4) was added to the reaction to supershift the RBP-J- RBPG3P1a complex.
To determine whether RBP-J binds to the (RG3P1a containing) upstream Gata3 promoter in vivo we performed chromatin precipitation. An antibody to RBP-J specifically precipitated the upstream promoter region from wild type CD4+ T cell derived chromatin, but not from chromatin obtained from RBP-J deficient CD4+ T cells (Figure 6a and b). The downstream gata3 promoter region did not precipitate with this antibody, consistent with the absence of RBP-J consensus sites in this region and its lack of responsiveness to Notch.
Figure 6. RBP-J binds the upstream Gata3 promoter in vivo.

(a) ChIP was performed on chromatin from CD4+ T cells using control antibody (white bars) or antibody to RBP-J (black bars). Precipitation of the upstream Gata3 promoter (USG3P) or the downstream Gata3 promoter (DSG3P) was determined by quantitative PCR. Samples were normalized to Input values. (b) ChIP was performed as in (a) using chromatin from wild type (black bars) or RBP-J deficient (white bars) CD4 T cells.
Gata3 is required for induction of Th2 differentiation by Notch
Although Gata3 is an important factor in differentiation of Th2 cells in response to IL4, its requirement in Notch induced Th2 differentiation has not been established. To test this, we studied Notch mediated Th2 differentiation in Gata3 deficient CD4 T cells. To circumvent effects from Gata3 deficiency on thymic development (Pai et al., 2003), we isolated mature CD4 T cells carrying floxed alleles of the gata3 gene and extinguished Gata3 expression by retroviral introduction of the Cre recombinase (Figure 7a). Expression of the Cre recombinase abrogated IL4 induced Th2 differentiation, documenting the efficacy of the deletion of the gata3 gene (Zhu et al., 2004) (and data not shown). To determine whether Gata3 is required for Notch induced Th2 differentiation, we carried out double infections of gata3flox/− CD4 T cells with Cre and NICD encoding retrovirus. To prevent potential induction of Gata3 expression by NICD before complete deletion of the gata3 gene, we performed these infections sequentially. Thus, cells were infected with Cre retrovirus first, and 6 hours later underwent a second round of infection with NICD. We minimized spontaneous effector differentiation by adding blocking antibodies to IL4 and IFNγ. Indeed, these antibodies worked effectively, since production of IL4 and IFNγ by cells infected with control retroviruses was undetectable (Figure 7b). Importantly, NICD elicited marked IL4 production in control cells, but not in cells lacking Gata3 expression (Figure 7b). Thus, Gata3 is instrumental in Notch mediated Th2 responses. Strikingly, instead of eliciting production of IL4, in the absence of Gata3 NICD strongly induced production of IFNγ (Figure 7b). This was actively induced by Notch as vector control cells lacking Gata3 expression did not default to Th1 differentiation.
Figure 7. Gata3 is necessary for Notch induced Th2 responses.

(a) Naïve CD4+ T cells from Gata3flox/− mice were activated with splenic APC and antibodies to CD3 and CD28 and infected with control GFP virus or virus encoding Cre linked through an IRES sequence to GFP. After 3 days, GFP+ cells were isolated by FACSorting and RNA was made. Relative abundance of Gata3 message was determined by real time PCR using a primer probe set that detects the floxed exon 4 of gata3. (b) Naïve CD4+ T cells from Gata3flox/− mice were activated as in (a) and infected after 20 hours first with control IRES-GFP (black bars) or Cre-IRES-GFP (gray bars). 6 hours later, cells were transduced with control IRES-Thy1.1 virus (left) or NICD-IRES-Thy1.1 (right). Neutralizing antibodies to IL4 and IFNγ (10μg/ml each) were added to the cultures at the time of the second transduction. 3 days after the last transduction cells were sorted for GFP and Thy1.1 expression and restimulated with plate bound anti CD3 (10μg/ml). 48 hour supernatants were tested for IL4 and IFNγ concentrations by ELISA. These results are representative of 3 independent experiments.
Discussion
Notch controls Th1, or Th2, or both?
The role of Notch in T helper cell differentiation has been controversial, with different groups showing apparently contradictory results. Using indirect approaches to interfere with Notch signaling some studies supported a role in Th1 and others a role in the opposite Th2 responses (Amsen et al., 2004; Maekawa et al., 2003; Minter et al., 2005; Tanigaki et al., 2004; Tu et al., 2005). Here, using both RBP-J deficient and Notch1/2 deficient T cells we establish that both downstream signaling as well as the Notch receptors themselves are required in Th2 responses. This role of Notch was evident under physiological conditions with extract of Schistosoma mansoni eggs as Th2 inducing adjuvant. Notch was not required in the classical in vitro paradigm for Th2 differentiation (Minter et al., 2005) (Fig2B), in which skewing is achieved by addition of a high concentration of IL4 and blocking antibodies to IFNγ. We think, however, that these results illustrate a limitation of the cytokine driven differentiation paradigm. While this approach has been very useful in delineating many pathways involved in T helper differentiation (and has been used extensively by ourselves for this purpose), it may obscure the physiological role of pathways functioning upstream of these cytokines or under conditions where such strong skewing conditions are not met. While exogenous IL4 may be present in vivo, clearly, its levels are insufficient to drive Th2 differentiation in the absence of Notch signaling, as evidenced by the profound defect in Th2 induction for instance in RBPJ deficient mice.
Do our findings rule out a role for Notch in Th1 differentiation? We think not. Of course, our experiments were not designed to rigorously investigate the involvement of Notch in Th1 responses, as SEA does not generally elicit strong Th1 responses (Pearce et al., 2004). We did examine the consequence of Notch1/2 deficiency in cytokine driven Th1 differentiation and failed to find a defect. However, it is possible that other Notchs (3 and 4) are sufficient for this response. Our negative data stand in contrast to compelling results showing profound inhibition of Th1 differentiation by gamma secretase inhibitors in vitro (Minter et al., 2005). Also, pronounced Th1 induction is obtained when CD4 T cells are stimulated with Delta ligands (Amsen et al., 2004; Maekawa et al., 2003) and an active Notch allele induced a strong Th1 response when the gata3 gene had been deleted (Figure 7b). We therefore still consider it likely that Notch has a role in Th1 responses, although it remains unclear under which conditions this pathway normally operates.
How the Notch pathway would regulate such opposite differentiation pathways as Th1 and Th2 is not understood, but our data suggest that the ability of Notch to drive either pathway is dependent on whether or not Notch can activate expression of Gata3 (Figure 7B). This might be determined by qualitatively or quantitatively different signaling induced by different ligands, and/or by surrounding signals.
Th2 specification by Notch: inductive signaling or lateral inhibition?
The Th1 and Th2 differentiation processes involve both positive feedback and cross inhibitory mechanisms. IFNγ and IL4 promote Th1 and Th2 differentiation, respectively, in an auto/paracrine fashion (Murphy and Reiner, 2002). At the same time, these factors induce inhibitory signals towards the opposite differentiation program (Murphy and Reiner, 2002). In this light, it was tempting to speculate that Notch, which controls other differentiation processes through lateral inhibition, promotes default Th2 differentiation by preventing the Th1 differentiation program.
However, a lateral inhibition model seems difficult to reconcile with our finding that Notch actively induces Th1 differentiation in the absence of Gata3 (Figure 7b). Furthermore, we have revealed a direct positive connection between Notch and induction of expression of Gata3, the master regulator of Th2 differentiation. A direct link also exists between Notch and the il4 gene: we showed previously that a 3′ enhancer of the il4 gene contains conserved Notch responsive RBP-J sites (Amsen et al., 2004), which was confirmed by others (Tanaka et al., 2006). These direct positive connections between Notch and the induction of expression of key Th2 genes strongly favor an inductive model for Notch mediated Th2 differentiation, rather than one based on lateral inhibition.
Notch and the regulation of gata3 gene expression
Gata3 is a critical factor for Th2 cell differentiation. Its expression is both necessary and sufficient for Th2 differentiation (Murphy and Reiner, 2002) (Zhu et al., 2004). Gata3 exerts its function by orchestrating a remodeling of the chromatin structure of the Th2 cytokine locus as well as by direct transcriptional control of several cytokine genes (Ansel et al., 2006).
Remarkably little is known about the pathways inducing expression of this factor in T cells. What little has been learned has come from cytokine driven in vitro differentiation systems, which may override the role of early signals. Regulation of gata3 expression is likely to be complex. Two promoters exist controlling the expression of transcripts with different first exons (Asnagli et al., 2002). The genomic region surrounding the gata3 gene contains many highly conserved noncoding regions (unpublished observations) and several distal enhancer elements have been described(Burch, 2005).
In CD4+ T cells, expression of the gata3 gene is responsive to TCR and IL4 receptor/STAT6 signaling (Murphy and Reiner, 2002). How signaling downstream of these connects to transcriptional activation of the gata3 gene has not been established. A good candidate in the TCR dependent pathway is NFκB. The role of NFκB in T helper differentiation is not entirely clear, possibly reflecting distinct functions for different NFκB family members (Corn et al., 2005), but expression of Gata3 is reduced in p50 null mice (Das et al., 2001). P50 may, perhaps in combination with Bcl3, activate transcription of the gata3 gene by binding to a site in the downstream gata3 promoter (Corn et al., 2005). It is not known whether, and if so how, p50 is activated specifically under conditions predisposing towards Th2 development. Interestingly, p50 activity is elevated by Notch signaling, which promotes its nuclear retention (Shin et al., 2006). This provides a possible additional mechanism for Notch to promote Th2 responses, independent from the mechanism identified in the present report. Finally, stable expression of Gata3 may be achieved by a positive feedback mechanism, in which Gata3 promotes its own expression (Ouyang et al., 2000). Again, the elements responsible for this have not been identified.
Notch and Gata factors: old friends
Connections between Notch and Gata factors are not limited to Gata3; they exist in various cell types and are conserved in phylogeny. For instance, Notch controls expression of the Drosophila Gata factor Serpent (Mandal et al., 2004). Expression of the gata2 gene in early hematopoietic progenitors is also directly controlled by Notch (de Pooter et al., 2006; Kumano et al., 2001; Robert-Moreno et al., 2005). Importantly, in common lymphoid progenitor cells Notch signaling induces expression of Gata3, which is required for commitment to the T cell lineage (Hoflinger et al., 2004; Taghon et al., 2005). It is tempting to speculate that this involves the mechanism identified here by us.
Notch and Gata factors are important regulators of differentiation throughout the metazoan kingdom. In many processes expression of the latter is connected to instructive differentiation signals received at the cell surface by Notch, a transcription factor and cell surface receptor at the same time. As we and an accompanying paper (Fang et al.,) demonstrate, in T helper cells this module has been adopted for the induction of Th2 type immunity.
Methods
Reagents and Antibodies
Anti-RBP-J (K0043) (Institute of Immunology Co., Japan), anti-CD3 (145-2C11), anti-CD28 (37.1), anti-IL4 (11B11), anti-IFNγ (XMG1.1), NK1.1 (HB101), anti-Thy1 (Y19) (all American Type Tissue Culture Collection, Manassas, Virginia). Recombinant mouse IL4 and IL2 were from Pharmingen, recombinant mouse IL12 a gift from Wyeth Research. Antibodies for FACS, cytokine and IgE ELISA from Pharmingen. Other isotype specific ELISA antibodies from Southern Biotechnology Associates, Inc., (cat. nr. 5300–04). ELISAs were developed with Horseradish Perozidase Avidin D (Vector Laboratories, Inc., Burlingame, CA) and SureBlue Peroxidase Substrate (KPL, Gaithersburg, MD).
Mice
Five- to eight-week-old B6 mice were purchased from The Jackson Laboratory (Bar Harbor, Maine) or NCI (Rockville, MD) and maintained in the Yale University Animal Resources Center. Bred in our colony under SPF conditions: Notch-1 flox (Radtke et al., 1999), Notch2 flox (McCright et al., 2006), RBP-J null (Oka et al., 1995), RBP-J flox (Tanigaki et al., 2002), CD4-Cre transgenic mice (Taconic), STAT6 null and AND TCR transgenic mice (Jackson laboratories). The floxed Gata3F allele was generated by crossing Gata3ex4GFP/+ mice (Grote et al., 2006) with the transgenic FLPe line (Rodriguez et al., 2000).
Vectors and Constructs
The hCRE-GFP-RV and GFP-RV vectors were generously provided by Dr. W. Paul. MSCV Thy1.1 and N1-MSCV-Thy1.1 (containing the entire intracellular tail of human Notch1 starting at amino acid 1748, first amino acids RKRRRQ) were described previously (Amsen et al., 2004). Expression constructs pCMX-RBP-J and pCMX-RBP-J R218H (Kato et al., 1997) were provided by Dr. T. Honjo.
In vitro CD4+ Tcell differentiation
Naïve CD44lowCD62LhighDX5−CD25− CD4+ T cells were purified from spleen and peripheral lymph nodes by positive selection using anti CD4 beads (Miltenyi, cat#130-049-201) followed by FACsorting. Cells were cultured in Bruff’s medium (10% FCS, penicillin, streptomycin and L-glutamine). 105 naïve CD4+ T cells were cultured with 2.5×106 irradiated (2000rad) B10.BR splenocytes obtained by collagenase treatment (Collagenase D, Roche), 10μg/ml of pigeon cytochrome C (Sigma, cat# C-4011) and 20μg/ml SEA. For Th1 and Th2 cultures, 2 × 105 naïve CD4+ cells were activated by 4 × 106 irradiated (2000 rad) T cell and NK cell depleted C57Bl/6 splenocytes with soluble anti CD3 and anti CD28 (1μg/ml each), 10U/ml IL2 and 3.5ng/ml IL12 and 10 μg/ml anti IL4 (11B11) (Th1 cultures) or 1000 U/ml IL4 and 10μg/ml anti IFNγ (Th2 cultures). After 5 days, viable cells were harvested by fycoll (LSMOL Lymphocyte Separation Medium, Cappel), restimulated at 1×105 cells per well (96 well flat bottom plate, Falcon) with plate bound anti-CD3 (10μg/ml). Cytokine concentrations (48 hr supernatants) were determined by ELISA. For intracellular cytokine staining viable effector cells were isolated by fycoll gradient, stimulated with PMA (50ng/ml) and ionomycin (0.5μM) and stained using the BD Cytofix/Cytoperm Plus kit (with Golgi Stop) (BD Pharmingen).
Retroviral transductions
Virus was made and transductions were performed as described (Amsen et al., 2004). For double infections, cells were first infected with Cre expressing virus and 6 hours later with NICD retrovirus. 3 days after transduction viable cells were isolated by fycoll. GFP-positive and/or Thy-1.1-positive were isolated by FACSorting.
Induction of anti SEA responses
Extracts from Schistosoma mansoni eggs were prepared as described (Boros and Warren, 1970). Water soluble fraction was injected intraperitoneally (50 μg) twice a week for 3 weeks. Following an additional two week rest period sera and spleens were collected. To measure T cell responses, CD4+ T cells were isolated from spleens and restimulated in vitro with C57Bl/6 splenocytes and 25μg/ml SEA. Supernatants were collected after 4 days and cytokine concentrations determined by ELISA.
EMSA
Double strands oligos containing the following sequences (with 5′ G overhang) were made: G3P1a: GACCTCTGATGTGCGGTT, mG3P1a: GACCTCTCTTGTGCGGTT, RE: GGGCACTGTGGGAACGGAA. 100ng of the double stranded oligos was labeled using Klenow (New England BioLabs) and γ32P dCTP (Perkin Elmer) and purified by column chromatography with Sephadex G-50 (GE-Healthcare-Amersham). 30,000 to 60,000cpms of labeled oligos (1–5ng) were incubated with 4μg nuclear extract from Th2 cells or whole-cell extract from transfected CHO cells. Extracts were prepared as described (Amsen et al., 2004). Reactions were done in the presence of 2μg of poly dI:dC in binding buffer (100mM Hepes pH7.6, 4mM EDTA, 2mM DTT, 20mM MgCl2, 300mM KCl, 40% Glycerol, supplemented with proteinase inhibitors). For supershift assays, 1μg of rat anti-RBP-J (K0043) or isotype control antibody (11B11) was added for 3 hours prior to addition of probe. Samples were resolved on 5% polyacrylamide gels and visualized using HyBlot CL autoradiography film (Denville Scientific Inc.).
RT-PCR and Quantitative PCR
RNA was made using Trizol (Invitrogen, Life Technologies) and further purified with RNeasyTM columns (Qiagen). RNA was transcribed into cDNA with Oligo(dT)12–18 Primers and SuperScript II RNase H- RT Kit as described in the manuals (Invitrogen).
| GATA3 exon1a-exon2 splice variant primers: | TGTGGGAGCGTCAGCAACAG and AGGGAGAGAGGAATCCGAG |
| GATA3 exon1b-exon2 splice variant primers: | GAGACTGAGAGAGCGAGACATAG and GGAATCCGAGTGTGACCAC |
Primers and probes for detection of total GATA3 and HPRT were described (Amsen et al., 2004). Quantitative PCR was performed for 40 cycles using 7500 Real Time PCR System (Applied Biosystems). Relative concentrations were determined on basis of standard curves of cDNA and normalized for HPRT contents using software provided by the manufacturer. HPRT and GATA3 probes, as well as Power SYBR Green PCR Master Mix were purchased from Applied Biosystems. Melt curves were run to ensure amplification of a single product.
Chromatin immunoprecipitation
Chromatin was prepared and precipitated using the ChIP kit from Upstate Cell Signaling (cat# 17–295) essentially as per manufacturers recommendations with minor modifications: preclearing was performed for 1 hour and after precipitation two washes with each of the wash buffers were carried out. Chromatin of 2 × 106 Th2 cells was precipitated per sample using polyclonal goat anti RBP-J from Santa Cruz Biotechnology (sc-8213) or control goat serum (sc-2028) and Salmon Sperm coated Protein G coupled agarose beads (Upstate Cell signaling, cat# 16-201). Precipitation of the gata3 promoters was measured by quantitative PCR using the following primers:
| upstream gata3 promoter 5′-AATGACACTGCCCTGTGGAATG. |
| upstream gata3 promoter 3′-CCGTGCCCATAGAACCTCTTATTG |
| downstream gata3 promoter 5′-ATTCCCTCCTGCCTGTCCC |
| downstream gata3 promoter 3′-CAACCCAAACCCGCTCCAG. |
Acknowledgments
We thank Wyeth Research for generously providing recombinant IL12, Dr. Tasuku Honjo for mice carrying floxed RBP-J alleles and RBP-J expression constructs, Dr. William Paul for the hCre retroviral expression vector. Furthermore we thank Drs. Babis Spilianakis, Ila Joshi and Barbara Osborne for technical advise, Drs. Julie Blander and Patrick E. Fields for critical reading of manuscript, Robert Westland for technical assistance and Fran Manzo for assistance with preparation of the manuscript. R.A.F. is an investigator of the Howard Hughes Medical Institute and D.A. was an associate of the Howard Hughes Medical Institute. He was also supported by the American Diabetes Association and an AMC fellowship. TG was supported by a grant from the NIH (NS036437). The authors have no conflicting financial interests.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbas AK, Murphy KM, Sher A. Functional diversity of helper T lymphocytes. Nature. 1996;383:787–793. doi: 10.1038/383787a0. [DOI] [PubMed] [Google Scholar]
- Amsen D, Blander JM, Lee GR, Tanigaki K, Honjo T, Flavell RA. Instruction of distinct CD4 T helper cell fates by different notch ligands on antigen-presenting cells. Cell. 2004;117:515–526. doi: 10.1016/s0092-8674(04)00451-9. [DOI] [PubMed] [Google Scholar]
- Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol. 2006;24:607–656. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Asnagli H, Afkarian M, Murphy KM. Cutting edge: Identification of an alternative GATA-3 promoter directing tissue-specific gene expression in mouse and human. J Immunol. 2002;168:4268–4271. doi: 10.4049/jimmunol.168.9.4268. [DOI] [PubMed] [Google Scholar]
- Bailey AM, Posakony JW. Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 1995;9:2609–2622. doi: 10.1101/gad.9.21.2609. [DOI] [PubMed] [Google Scholar]
- Barolo S, Walker RG, Polyanovsky AD, Freschi G, Keil T, Posakony JW. A notch-independent activity of suppressor of hairless is required for normal mechanoreceptor physiology. Cell. 2000;103:957–969. doi: 10.1016/s0092-8674(00)00198-7. [DOI] [PubMed] [Google Scholar]
- Boros DL, Warren KS. Delayed hypersensitivity-type granuloma formation and dermal reaction induced and elicited by a soluble factor isolated from Schistosoma mansoni eggs. J Exp Med. 1970;132:488–507. doi: 10.1084/jem.132.3.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- Burch JB. Regulation of GATA gene expression during vertebrate development. Semin Cell Dev Biol. 2005;16:71–81. doi: 10.1016/j.semcdb.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Corn RA, Hunter C, Liou HC, Siebenlist U, Boothby MR. Opposing roles for RelB and Bcl-3 in regulation of T-box expressed in T cells, GATA-3, and Th effector differentiation. J Immunol. 2005;175:2102–2110. doi: 10.4049/jimmunol.175.4.2102. [DOI] [PubMed] [Google Scholar]
- Das J, Chen CH, Yang L, Cohn L, Ray P, Ray A. A critical role for NF-kappa B in GATA3 expression and TH2 differentiation in allergic airway inflammation. Nat Immunol. 2001;2:45–50. doi: 10.1038/83158. [DOI] [PubMed] [Google Scholar]
- de La Coste A, Freitas AA. Notch signaling: distinct ligands induce specific signals during lymphocyte development and maturation. Immunol Lett. 2006;102:1–9. doi: 10.1016/j.imlet.2005.06.014. [DOI] [PubMed] [Google Scholar]
- de Pooter RF, Schmitt TM, de la Pompa JL, Fujiwara Y, Orkin SH, Zuniga-Pflucker JC. Notch signaling requires GATA-2 to inhibit myelopoiesis from embryonic stem cells and primary hemopoietic progenitors. J Immunol. 2006;176:5267–5275. doi: 10.4049/jimmunol.176.9.5267. [DOI] [PubMed] [Google Scholar]
- Fallon PG, Richardson EJ, McKenzie GJ, McKenzie AN. Schistosome infection of transgenic mice defines distinct and contrasting pathogenic roles for IL-4 and IL-13: IL-13 is a profibrotic agent. J Immunol. 2000;164:2585–2591. doi: 10.4049/jimmunol.164.5.2585. [DOI] [PubMed] [Google Scholar]
- Fang, etc Pear paper
- Finkelman FD, Morris SC, Orekhova T, Mori M, Donaldson D, Reiner SL, Reilly NL, Schopf L, Urban JF., Jr Stat6 regulation of in vivo IL-4 responses. J Immunol. 2000;164:2303–2310. doi: 10.4049/jimmunol.164.5.2303. [DOI] [PubMed] [Google Scholar]
- Flynn S, Toellner KM, Raykundalia C, Goodall M, Lane P. CD4 T cell cytokine differentiation: the B cell activation molecule, OX40 ligand, instructs CD4 T cells to express interleukin 4 and upregulates expression of the chemokine receptor, Blr-1. J Exp Med. 1998;188:297–304. doi: 10.1084/jem.188.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote D, Souabni A, Busslinger M, Bouchard M. Pax 2/8-regulated Gata 3 expression is necessary for morphogenesis and guidance of the nephric duct in the developing kidney. Development. 2006;133:53–61. doi: 10.1242/dev.02184. [DOI] [PubMed] [Google Scholar]
- Hoflinger S, Kesavan K, Fuxa M, Hutter C, Heavey B, Radtke F, Busslinger M. Analysis of Notch1 function by in vitro T cell differentiation of Pax5 mutant lymphoid progenitors. J Immunol. 2004;173:3935–3944. doi: 10.4049/jimmunol.173.6.3935. [DOI] [PubMed] [Google Scholar]
- Hwang ES, Szabo SJ, Schwartzberg PL, Glimcher LH. T helper cell fate specified by kinase-mediated interaction of T-bet with GATA-3. Science. 2005;307:430–433. doi: 10.1126/science.1103336. [DOI] [PubMed] [Google Scholar]
- Ito T, Wang YH, Duramad O, Hori T, Delespesse GJ, Watanabe N, Qin FX, Yao Z, Cao W, Liu YJ. TSLP-activated dendritic cells induce an inflammatory T helper type 2 cell response through OX40 ligand. J Exp Med. 2005;202:1213–1223. doi: 10.1084/jem.20051135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankovic D, Kullberg MC, Noben-Trauth N, Caspar P, Paul WE, Sher A. Single cell analysis reveals that IL-4 receptor/Stat6 signaling is not required for the in vivo or in vitro development of CD4+ lymphocytes with a Th2 cytokine profile. J Immunol. 2000;164:3047–3055. doi: 10.4049/jimmunol.164.6.3047. [DOI] [PubMed] [Google Scholar]
- Kapsenberg ML. Dendritic-cell control of pathogen-driven T-cell polarization. Nat Rev Immunol. 2003;3:984–993. doi: 10.1038/nri1246. [DOI] [PubMed] [Google Scholar]
- Katada T, Ito M, Kojima Y, Miyatani S, Kinoshita T. XMam1, Xenopus Mastermind1, induces neural gene expression in a Notch-independent manner. Mech Dev. 2006 doi: 10.1016/j.mod.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Kato H, Taniguchi Y, Kurooka H, Minoguchi S, Sakai T, Nomura-Okazaki S, Tamura K, Honjo T. Involvement of RBP-J in biological functions of mouse Notch1 and its derivatives. Development. 1997;124:4133–4141. doi: 10.1242/dev.124.20.4133. [DOI] [PubMed] [Google Scholar]
- Kumano K, Chiba S, Shimizu K, Yamagata T, Hosoya N, Saito T, Takahashi T, Hamada Y, Hirai H. Notch1 inhibits differentiation of hematopoietic cells by sustaining GATA-2 expression. Blood. 2001;98:3283–3289. doi: 10.1182/blood.v98.12.3283. [DOI] [PubMed] [Google Scholar]
- Kurata H, Lee HJ, O’Garra A, Arai N. Ectopic expression of activated Stat6 induces the expression of Th2-specific cytokines and transcription factors in developing Th1 cells. Immunity. 1999;11:677–688. doi: 10.1016/s1074-7613(00)80142-9. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Takemoto N, Kurata H, Kamogawa Y, Miyatake S, O’Garra A, Arai N. GATA-3 induces T helper cell type 2 (Th2) cytokine expression and chromatin remodeling in committed Th1 cells. J Exp Med. 2000;192:105–115. doi: 10.1084/jem.192.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa Y, Tsukumo S, Chiba S, Hirai H, Hayashi Y, Okada H, Kishihara K, Yasutomo K. Delta1-Notch3 interactions bias the functional differentiation of activated CD4+ T cells. Immunity. 2003;19:549–559. doi: 10.1016/s1074-7613(03)00270-x. [DOI] [PubMed] [Google Scholar]
- Maillard I, Fang T, Pear WS. Regulation of lymphoid development, differentiation, and function by the Notch pathway. Annu Rev Immunol. 2005;23:945–974. doi: 10.1146/annurev.immunol.23.021704.115747. [DOI] [PubMed] [Google Scholar]
- Mandal L, Banerjee U, Hartenstein V. Evidence for a fruit fly hemangioblast and similarities between lymph-gland hematopoiesis in fruit fly and mammal aorta-gonadal-mesonephros mesoderm. Nat Genet. 2004;36:1019–1023. doi: 10.1038/ng1404. [DOI] [PubMed] [Google Scholar]
- McCright B, Lozier J, Gridley T. Generation of new Notch2 mutant alleles. Genesis. 2006;44:29–33. doi: 10.1002/gene.20181. [DOI] [PubMed] [Google Scholar]
- Minter LM, Turley DM, Das P, Shin HM, Joshi I, Lawlor RG, Cho OH, Palaga T, Gottipati S, Telfer JC, et al. Inhibitors of gamma-secretase block in vivo and in vitro T helper type 1 polarization by preventing Notch upregulation of Tbx21. Nat Immunol. 2005;6:680–688. [PubMed] [Google Scholar]
- Moser M, Murphy KM. Dendritic cell regulation of TH1–TH2 development. Nat Immunol. 2000;1:199–205. doi: 10.1038/79734. [DOI] [PubMed] [Google Scholar]
- Mosmann TR, Coffman RL. TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu Rev Immunol. 1989;7:145–173. doi: 10.1146/annurev.iy.07.040189.001045. [DOI] [PubMed] [Google Scholar]
- Mowen KA, Glimcher LH. Signaling pathways in Th2 development. Immunol Rev. 2004;202:203–222. doi: 10.1111/j.0105-2896.2004.00209.x. [DOI] [PubMed] [Google Scholar]
- Mullen AC, High FA, Hutchins AS, Lee HW, Villarino AV, Livingston DM, Kung AL, Cereb N, Yao TP, Yang SY, Reiner SL. Role of T-bet in commitment of TH1 cells before IL-12-dependent selection. Science. 2001;292:1907–1910. doi: 10.1126/science.1059835. [DOI] [PubMed] [Google Scholar]
- Murphy KM, Reiner SL. The lineage decisions of helper T cells. Nat Rev Immunol. 2002;2:933–944. doi: 10.1038/nri954. [DOI] [PubMed] [Google Scholar]
- Noben-Trauth N, Shultz LD, Brombacher F, Urban JF, Jr, Gu H, Paul WE. An interleukin 4 (IL-4)-independent pathway for CD4+ T cell IL-4 production is revealed in IL-4 receptor-deficient mice. Proc Natl Acad Sci U S A. 1997;94:10838–10843. doi: 10.1073/pnas.94.20.10838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka C, Nakano T, Wakeham A, de la Pompa JL, Mori C, Sakai T, Okazaki S, Kawaichi M, Shiota K, Mak TW, Honjo T. Disruption of the mouse RBP-J kappa gene results in early embryonic death. Development. 1995;121:3291–3301. doi: 10.1242/dev.121.10.3291. [DOI] [PubMed] [Google Scholar]
- Ouyang W, Lohning M, Gao Z, Assenmacher M, Ranganath S, Radbruch A, Murphy KM. Stat6-independent GATA-3 autoactivation directs IL-4-independent Th2 development and commitment. Immunity. 2000;12:27–37. doi: 10.1016/s1074-7613(00)80156-9. [DOI] [PubMed] [Google Scholar]
- Ouyang W, Ranganath SH, Weindel K, Bhattacharya D, Murphy TL, Sha WC, Murphy KM. Inhibition of Th1 development mediated by GATA-3 through an IL-4-independent mechanism. Immunity. 1998;9:745–755. doi: 10.1016/s1074-7613(00)80671-8. [DOI] [PubMed] [Google Scholar]
- Pai SY, Truitt ML, Ho IC. GATA-3 deficiency abrogates the development and maintenance of T helper type 2 cells. Proc Natl Acad Sci U S A. 2004;101:1993–1998. doi: 10.1073/pnas.0308697100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai SY, Truitt ML, Ting CN, Leiden JM, Glimcher LH, Ho IC. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity. 2003;19:863–875. doi: 10.1016/s1074-7613(03)00328-5. [DOI] [PubMed] [Google Scholar]
- Pearce EJ, C MK, Sun J, J JT, McKee AS, Cervi L. Th2 response polarization during infection with the helminth parasite Schistosoma mansoni. Immunol Rev. 2004;201:117–126. doi: 10.1111/j.0105-2896.2004.00187.x. [DOI] [PubMed] [Google Scholar]
- Radtke F, Wilson A, Stark G, Bauer M, van Meerwijk J, MacDonald HR, Aguet M. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10:547–558. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- Rincon M, Anguita J, Nakamura T, Fikrig E, Flavell RA. Interleukin (IL)-6 directs the differentiation of IL-4-producing CD4+ T cells. J Exp Med. 1997;185:461–469. doi: 10.1084/jem.185.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Moreno A, Espinosa L, de la Pompa JL, Bigas A. RBPjkappa-dependent Notch function regulates Gata2 and is essential for the formation of intra-embryonic hematopoietic cells. Development. 2005;132:1117–1126. doi: 10.1242/dev.01660. [DOI] [PubMed] [Google Scholar]
- Schmitz J, Thiel A, Kuhn R, Rajewsky K, Muller W, Assenmacher M, Radbruch A. Induction of interleukin 4 (IL-4) expression in T helper (Th) cells is not dependent on IL-4 from non-Th cells. J Exp Med. 1994;179:1349–1353. doi: 10.1084/jem.179.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, McElhinny AS, Cao Y, Gao P, Liu J, Bronson R, Griffin JD, Wu L. The Notch coactivator, MAML1, functions as a novel coactivator for MEF2C-mediated transcription and is required for normal myogenesis. Genes Dev. 2006;20:675–688. doi: 10.1101/gad.1383706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin HM, Minter LM, Cho OH, Gottipati S, Fauq AH, Golde TE, Sonenshein GE, Osborne BA. Notch1 augments NF-kappaB activity by facilitating its nuclear retention. Embo J. 2006;25:129–138. doi: 10.1038/sj.emboj.7600902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So T, Song J, Sugie K, Altman A, Croft M. Signals from OX40 regulate nuclear factor of activated T cells c1 and T cell helper 2 lineage commitment. Proc Natl Acad Sci U S A. 2006;103:3740–3745. doi: 10.1073/pnas.0600205103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- Tacchini-Cottier F, Allenbach C, Otten LA, Radtke F. Notch1 expression on T cells is not required for CD4+ T helper differentiation. Eur J Immunol. 2004;34:1588–1596. doi: 10.1002/eji.200324337. [DOI] [PubMed] [Google Scholar]
- Taghon TN, David ES, Zuniga-Pflucker JC, Rothenberg EV. Delayed, asynchronous, and reversible T-lineage specification induced by Notch/Delta signaling. Genes Dev. 2005;19:965–978. doi: 10.1101/gad.1298305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Tsukada J, Suzuki W, Hayashi K, Tanigaki K, Tsuji M, Inoue H, Honjo T, Kubo M. The interleukin-4 enhancer CNS-2 is regulated by Notch signals and controls initial expression in NKT cells and memory-type CD4 T cells. Immunity. 2006;24:689–701. doi: 10.1016/j.immuni.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Tanigaki K, Han H, Yamamoto N, Tashiro K, Ikegawa M, Kuroda K, Suzuki A, Nakano T, Honjo T. Notch-RBP-J signaling is involved in cell fate determination of marginal zone B cells. Nat Immunol. 2002;3:443–450. doi: 10.1038/ni793. [DOI] [PubMed] [Google Scholar]
- Tanigaki K, Tsuji M, Yamamoto N, Han H, Tsukada J, Inoue H, Kubo M, Honjo T. Regulation of alphabeta/gammadelta T cell lineage commitment and peripheral T cell responses by Notch/RBP-J signaling. Immunity. 2004;20:611–622. doi: 10.1016/s1074-7613(04)00109-8. [DOI] [PubMed] [Google Scholar]
- Tu L, Fang TC, Artis D, Shestova O, Pross SE, Maillard I, Pear WS. Notch signaling is an important regulator of type 2 immunity. J Exp Med. 2005;202:1037–1042. doi: 10.1084/jem.20050923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tun T, Hamaguchi Y, Matsunami N, Furukawa T, Honjo T, Kawaichi M. Recognition sequence of a highly conserved DNA binding protein RBP-J kappa. Nucleic Acids Res. 1994;22:965–971. doi: 10.1093/nar/22.6.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui T, Nishikomori R, Kitani A, Strober W. GATA-3 suppresses Th1 development by downregulation of Stat4 and not through effects on IL-12Rbeta2 chain or T-bet. Immunity. 2003;18:415–428. doi: 10.1016/s1074-7613(03)00057-8. [DOI] [PubMed] [Google Scholar]
- Usui T, Preiss JC, Kanno Y, Yao ZJ, Bream JH, O’Shea JJ, Strober W. T-bet regulates Th1 responses through essential effects on GATA-3 function rather than on IFNG gene acetylation and transcription. J Exp Med. 2006;203:755–766. doi: 10.1084/jem.20052165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver CT, Hatton RD, Mangan PR, Harrington LE. IL-17 Family Cytokines and the Expanding Diversity of Effector T Cell Lineages. Annu Rev Immunol. 2007 doi: 10.1146/annurev.immunol.25.022106.141557. [DOI] [PubMed] [Google Scholar]
- Wolfe MS, Kopan R. Intramembrane proteolysis: theme and variations. Science. 2004;305:1119–1123. doi: 10.1126/science.1096187. [DOI] [PubMed] [Google Scholar]
- Yamashita M, Ukai-Tadenuma M, Miyamoto T, Sugaya K, Hosokawa H, Hasegawa A, Kimura M, Taniguchi M, DeGregori J, Nakayama T. Essential role of GATA3 for the maintenance of type 2 helper T (Th2) cytokine production and chromatin remodeling at the Th2 cytokine gene loci. J Biol Chem. 2004;279:26983–26990. doi: 10.1074/jbc.M403688200. [DOI] [PubMed] [Google Scholar]
- Zhang DH, Cohn L, Ray P, Bottomly K, Ray A. Transcription factor GATA-3 is differentially expressed in murine Th1 and Th2 cells and controls Th2-specific expression of the interleukin-5 gene. J Biol Chem. 1997;272:21597–21603. doi: 10.1074/jbc.272.34.21597. [DOI] [PubMed] [Google Scholar]
- Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- Zhu J, Min B, Hu-Li J, Watson CJ, Grinberg A, Wang Q, Killeen N, Urban JF, Jr, Guo L, Paul WE. Conditional deletion of Gata3 shows its essential function in T(H)1-T(H)2 responses. Nat Immunol. 2004;5:1157–1165. doi: 10.1038/ni1128. [DOI] [PubMed] [Google Scholar]