Abstract
Previously we showed that overexpression of ubiquilin reduces protein aggregates and toxicity of expanded polyglutamine proteins. Here, we investigated the mechanism of ubiquilin's protective effect. Immunofluorescence microscopy and immunoprecipitation studies indicated that ubiquilin colocalized and coimmunoprecipitated more with GFP-huntingtin-exon-1-fusion proteins containing a 74-polyglutamine tract than with GFP-huntingtin-fusion proteins containing a 28-polyglutamine tract or with GFP protein alone. Furthermore, overexpression of ubiquilin selectively enhanced the turnover of the expanded GFP-huntingtin-fusion protein. These results suggest that elevating ubiquilin levels could aid in the selective disposal of potentially toxic expanded polyglutamine proteins that are thought to cause several human diseases.
Keywords: Huntington's disease, polyglutamine, ubiquilin, aggregation, protein turnover
Introduction
Huntington's disease (HD) is an autosomal dominant neurodegenerative disease caused by abnormal expansion of a CAG trinucleotide repeat in the first exon of the huntingtin (htt) gene [1]. The number of CAG repeats in htt is the key factor that predicts whether a person will develop HD. CAG repeats numbering 34 or fewer do not cause HD, whereas repeats numbering between 35 to 39 show incomplete penetrance, whereas repeats numbering 40 or more displaying full penetrance [2-4]. It is now well established that the longer the number of CAG repeats the earlier the age of onset of HD [3, 5]. The CAG repeats in htt are translated into a reiterated stretch of glutamine amino acids. In fact, HD is one of nine neurological disorders that are associated with an expansion of polyglutamine tracts in otherwise unrelated proteins [4]. The exact mechanism by which the expanded polyglutamine proteins cause disease is still debated although a number of studies suggest that it is related to a toxic gain-of-function, possibly related to increased propensity of the expanded polyglutamine proteins to misfold and aggregate [2, 6].
A number of polyglutamine aggregate-interacting proteins have been identified, one of which is ubiquilin [7]. Ubiquilin was first identified as a presenilin-interacting protein and is classified as a type-2 ubiquitin-like protein [8]. Type-2 ubiquitin-like proteins, unlike the type-1 proteins, are not covalently attached onto target proteins [9]. Ubiquilin contains three domains: an N-terminal ubiquitin-like (UBL) domain, a central variable region, and a C-terminal ubiquitin-associated (UBA) domain. There are three ubiquilin isoforms expressed in mammals: ubiquilin-1 is expressed in all tissues examined [8, 10], ubiquilin-2 has a more restricted tissue expression pattern than ubiquilin-1 [10], whereas ubiquilin-3 is expressed only in the testis [11]. Evolutionarily, ubiquilin proteins are found in all species examined suggesting it serves some important function. One speculation is that ubiquilin functions as a shuttle factor to deliver misfolded proteins to the proteasome for degradation. This property would be in accord with the ability of ubiquilin proteins to bind ubiquitin moieties conjugated onto proteins via its UBA domain and subunits of the proteasome via its UBL domain [12-14].
In a previous report we showed that overexpression of ubiquilin suppresses the formation of protein aggregates and toxicity of expanded polyglutamine proteins [15]. Here, we have examined how ubiquilin might exert this protective effect. We demonstrate that ubiquilin not only binds selectively to expanded polyglutamine proteins, but it also enhances their degradation.
Materials and Methods
Polyglutamine stable cell lines, cell culture, and drug treatment
The generation of HeLa cell lines stably expressing GFP, or GFP-htt-Exon1-28Q, or GFP-htt-74Q fusion proteins (referred to as GFP, GFP-28Q and GFP-74Q cell lines, respectively) were described previously [15]. The stable cell lines were cultured in DMEM supplemented with 10% FBS and 0.1% (g/m1) of G418 (Invitrogen).
Plasmids and DNA transfection
The ubiquilin-1 cDNA expression plasmid used here has been described previously [8] [14]. The GFP-28Q and GFP-74Q cells were transiently transfected with a ubiquilin-1 cDNA expression plasmid or mock transfected (control) using the calcium phosphate co-precipitation method.
Immunoprecipitation and immunoblotting
GFP, GFP-28Q, and GFP-74Q cell cultures grown in 10 cm dishes were washed with ice-cold PBS and the cells harvested by scraping in immunoprecipitation buffer (IP: 50 mM Tris, pH 7.50, 150 mM NaCl, 2 mM EDTA, 0.5% NP-40, and protease inhibitor cocktails). The cells were then disrupted by gently forcing the cell suspension through a 25-gage needle, eight times. The lysate was centrifuged at 5000 g for 5 min and resulting supernatant was saved, and the pellet discarded. Supernatants, containing equivalent amounts of protein from each cell line, were incubated with a GFP polyclonal antibody and protein-A Sepharose beads for 2 hours at 4 C with gentle rotation. The mixtures were then centrifuged to recover the immunoprecipitates, which were then washed with IP buffer and finally resuspended in SDS-sample buffer. The samples were then boiled for 10 minutes, centrifuged briefly, and equivalent amounts of the supernatants separated by SDS-PAGE. The proteins were transferred to nitrocellullose membranes, and immunoblotted with either a mouse monoclonal anti-ubiquilin antibody (Zymed), a rabbit-GFP polyclonal antibody (described previously [15]), or with a monoclonal anti-ubiquitin antibody (Santa-Cruz Biotechnology).
Pulse-chase experiments
The rate of turnover of GFP-htt-28Q and -74Q fusion proteins were examined by classical 35S-methionine pulse-chase studies using essentially the same procedure as described previously [8]. Briefly, GFP-28Q and GFP-74Q cell cultures that had either been transfected with the ubiquilin-1 expression plasmid, or which were mock transfected, were pulse-labeled for 1 h with 150 μCi of 35S-methionine and then chased with non-radioactive culture medium for the times periods as indicated in the figures. The cells were then lysed and GFP and ubiquilin proteins were immunoprecipitated from equal portions of the lysates by addition of rabbit anti-GFP or anti-ubiquilin antibodies using our standard immunoprecipitation protocol. The immunoprecipitated-proteins were separated by SDS-PAGE, after which the gels were dried and exposed to a phosphoimage screen. The 35S-methionine radioactivity of appropriate bands was quantified after scanning the screens using a Typhoon Phosphoimage System (Amersham Bioscience) using ImageQuant software (Amersham Bioscience).
Results
Ubiquilin interacts more with GFP-htt-fusion proteins containing a long, rather than a short, polyglutamine tract
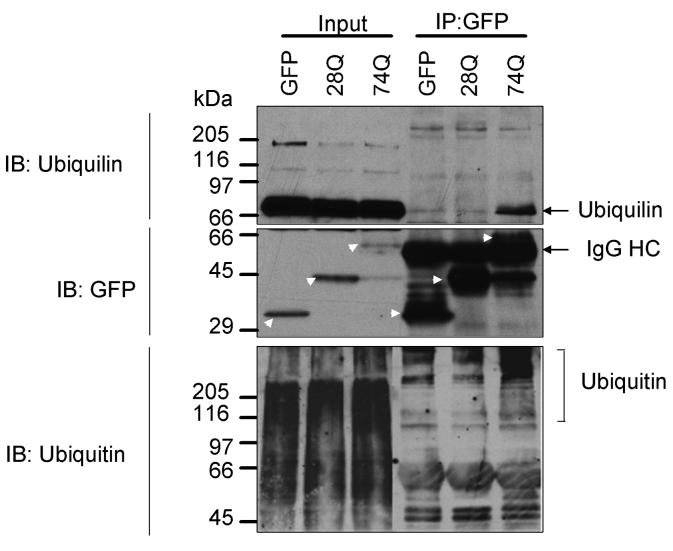
To investigate how ubiquilin modulates aggregation and toxicity of expanded polyglutamine proteins we first examined whether the proteins interact with each other in cells by double immunofluorescence microscopy. Confocal microscopy revealed that endogenous ubiquilin staining was colocalized with GFP-containing aggregates seen in the GFP-74Q cell line (Fig. 1A). Furthermore, transfection of the GFP-74Q cell line with a plasmid encoding ubiquilin-1 tagged with a red-fluorescent protein (mRFP) reporter led to colocalization of the red fluorescent signal with GFP-containing aggregates in the cells (Fig. 1B). We did not observe similar colocalization of ubiquilin proteins in the GFP-28Q cell line, in which GFP-protein aggregates are not visible (data not shown). This phenomenon led us to speculate that ubiquilin might interact only with aggregated GFP-polyglutamine proteins. Because the number of polyglutamine repeats influences polyglutamine protein aggregation we tested our hypothesis by examining whether ubiquilin interacts more strongly with GFP proteins containing longer polyglutamine repeats. To analyze this, we immunoprecipitated GFP proteins from cell lysates derived from the GFP, GFP-28Q, or GFP-74Q cell lines and examined the precipitates for coimmunoprecipitation of ubiquilin by immunoblotting. As seen in Figure 2 (upper panel), more ubiquilin was co-immunoprecipitated with GFP-74Q than with GFP-28Q or GFP proteins. Because ubiquilin binds ubiquitin moieties that are attached onto proteins [14, 16, 17] we speculated that the increase in ubiquilin binding to the GFP-htt-74Q protein could be due to increased ubiquitination of the GFP-htt-74Q protein compared to the GFP-htt-28Q and GFP proteins. Indeed an immunoblot of the immunoprecipitated GFP proteins revealed stronger anti-ubiquitin immunoactivity with proteins isolated from the cell line expressing GFP-htt-74Q than from lines expressing GFP-htt-28Q or GFP alone (Fig 2 bottom panel). In fact, the increased anti-ubiquitin immunoreactivity was mainly with proteins that migrated as a smear in the high molecular mass region of the gel, consistent with heavy polyubiquitination of the aggregate prone GFP-htt-74Q protein. These results indicate that ubiquilin interacts with expanded GFP-polyglutamine fusion proteins that are more likely to be polyubiquitinated.
Fig. 1. Ubiquilin colocalizes with GFP-htt-74Q aggregates.
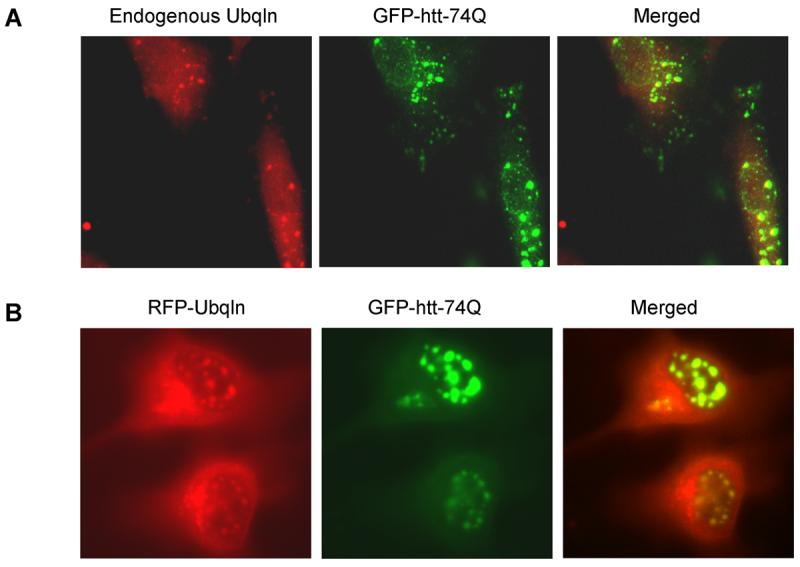
- Endogenous ubiquilin is colocalized with GFP-htt-74Q aggregates. GFP-74 cells were fixed, immunostained with a ubiquilin monoclonal antibody, and confocal microscopy was performed. Left panels (green), GFP-htt-74Q; center panels (red), endogenous ubiquilin; right panels, result of merging the green and red images.
- Transiently transfected RFP-ubiquilin-1 is colocalized with GFP-htt-74Q aggregates. GFP-74 cells were transiently transfected with a RFP-ubiquilin-1 expression construct and after 24 hr, confocal microscopy was performed without any antibody staining. Left panels (green), GFP-htt-74Q; center panels (red), RFP-ubiquilin; right panels, result of merging the green and red images.
Fig. 2. Ubiquilin interacts with GFP-htt-74Q proteins.
Coimmunoprecipitation of endogenous ubiquilin with normal and expanded polyglutamine-fusion proteins. Cells stably expressing GFP, GFP-htt-28Q, or GFP-htt-74Q were lysed and the GFP proteins were immunoprecipitated with a rabbit polyclonal GFP antibody. Equal portions of a representative portion of the lysates and immunoprecipitates were then separated by SDS-PAGE, the proteins were transferred to a nitrocellulose membrane, and then immunoblotted with either a monoclonal ubiquilin antibody (upper panel), GFP antibody (middle panel), or monoclonal ubiquitin antibody (lower panel). The position of the GFP proteins are indicated by the arrow head, but some of these bands in the immunoprecipitate lanes are obscured by cross-reaction of the GFP antibody with the IgG heavy chain of the antibody used in the immunoprecipitation.
Overexpression of ubiquilin selectively enhances the turnover of expanded GFP-httpolyglutamine-fusion proteins
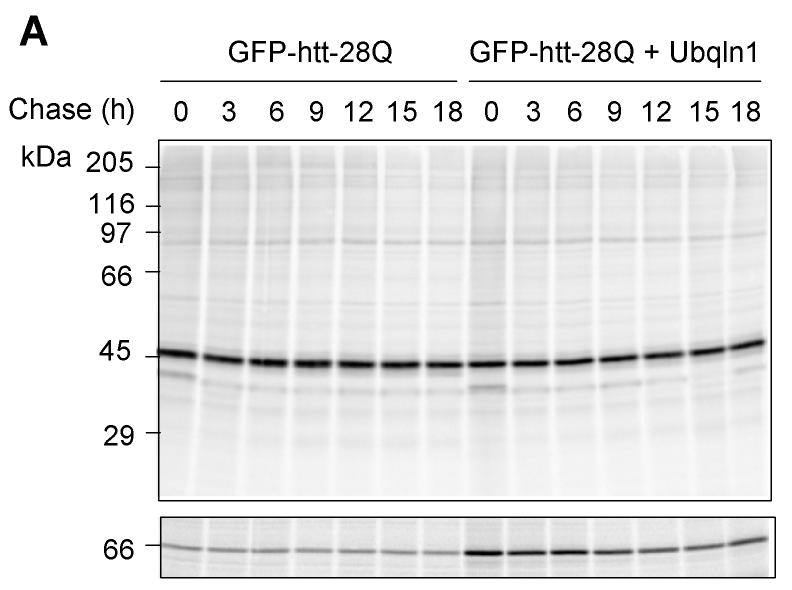
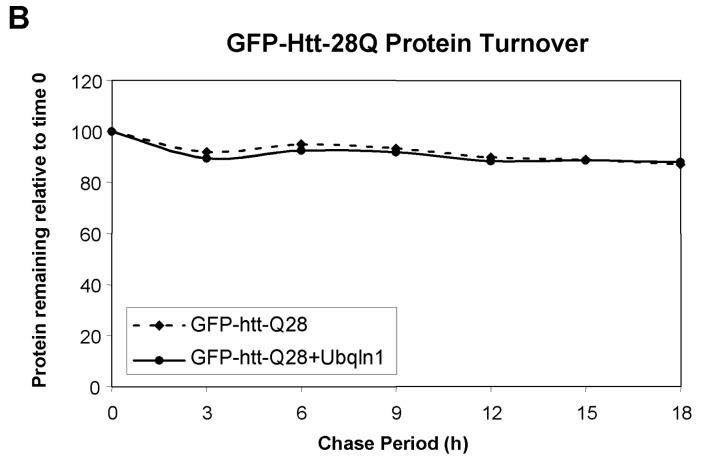
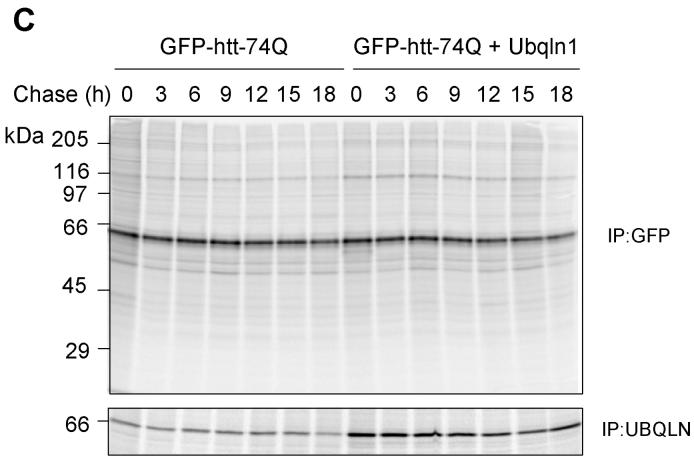
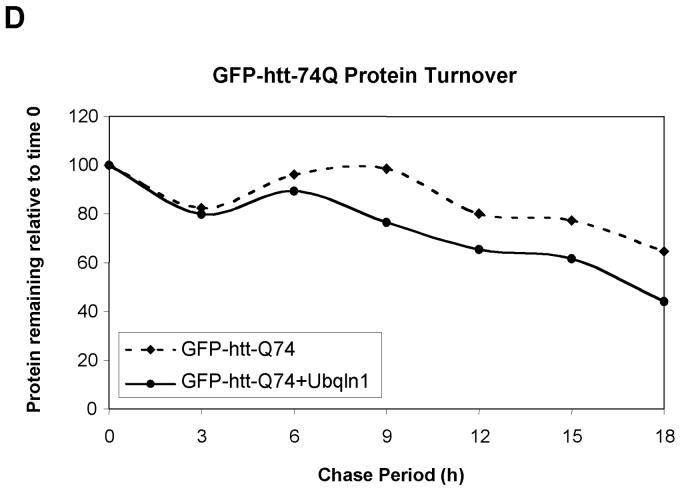
Because ubiquilin is thought to act as a shuttle factor that delivers ubiquitin-tagged proteins to the proteasome for degradation, we considered that its differential binding to GFP-httpolyglutamine proteins containing normal and expanded polyglutamine tracts might affect the turnover of the proteins. To analyze this, we conducted classical 35S-methionine pulse-chase studies examining the rate of turnover of GFP-htt-74Q and GFP-htt-28Q proteins in cells that had been transfected with or without a ubiquilin-1 expression construct. As shown in Figure 3B and 3D, the turnover of GFP-htt-74Q was enhanced in the cells transfected with the ubiquilin-1 expression construct compared to the mock-transfected cells. For example, 9 hr after labeling, the amount of radioactive immunoprecipitated GFP-htt-Q74 protein remaining in cells transfected with ubiquilin-1 was approximately 20% lower than in the mock-transfected cells, and this difference persisted for the next 9-hour duration of the experiment (Fig. 3D). A parallel immunoprecipitation of ubiquilin proteins in the two experimental conditions revealed that ubiquilin was indeed overexpressed at least two-fold higher in the cells transfected with ubiquilin-1 compared to mock-transfected cells (Fig. 3C, lower panel).
Fig. 3. Overexpression of ubiquilin-1 enhances the degradation of expanded GFP-htt-74Q protein.
- GFP-28Q cell line was either mockingly transfected (control) or transfected with a ubiquilin-1 expression plasmid (+ Ubiquilin-1). The cells were pulse-labeled with 35S-methionine for 1 h and then chased in non-radioactive medium for different times. At appropriate time intervals, as indicated in the figure, sets of cultures were lysed and equal fractions of the lysates were used to immunoprecipitate GFP or ubiquilin proteins. The immunoprecipitated proteins were separated by SDS-PAGE and then subjected to autoradiography. Autoradiography of GFP-htt-28Q (top panel) or ubiquilin (bottom panel) in mock (left half) or ubiquilin-1 transfected cells (right half).
- Quantitative measurement of the percent of radiolabeled GFP-htt-28Q protein recovered at different time points compared to the 0 h time point in the mock and ubiquilin-1 transfected cells in the experiment shown in A above.
- Autoradiograph of the immunoprecipitation of GFP-htt-74Q (top panel) or ubiquilin (bottom panel) proteins from the GFP-74Q cell line using the same protocol as described in Figure A.
- Quantitative measurement of the percent of radiolabeled GFP-htt-74Q protein recovered at different time points compared to the 0 h time point in the mock and ubiquilin-1 transfected cells in the experiment shown in Figure C.
By contrast, the rate of turnover of GFP-htt-28Q did not differ between cells that were transfected with ubiquilin-1 and those that were mock-transfected (Fig. 3A and 3C). Interestingly however, an examination of the turnover of GFP-htt-28Q and GFP-htt-Q74 proteins in the mock-transfected cells revealed that the former turned over more slowly than the latter, which we speculate is due to increased propensity of the latter to aggregate, be ubiquitinated, thereby increasing its likelihood to interact with endogenous ubiquilin and to be escorted for degradation. Because overexpression of ubiquilin enhanced the turnover of the GFP-htt-Q74 protein, our results suggests that under normal conditions the amount of endogenous ubiquilin capable of interacting with the GFP-htt-Q74 protein may be limiting in cells. Taken together, these results indicate that overexpression of ubiquilin selectively enhances the turnover of proteins containing an expanded polyglutamine tract, without affecting the turnover of GFP-htt-proteins containing a normal polyglutamine tract.
Discussion
Here, we present evidence showing ubiquilin binds more with GFP-htt-polyglutamine fusion proteins containing a stretch of 74 glutamine residues, in the range of polyglutamine repeats associated with causing toxicity, than with GFP-htt-fusion proteins containing 28 glutamine residues, a number not associated with toxicity. Moreover, we found that overexpression of ubiquilin-1 enhances the turnover of GFP-htt-74Q proteins but does not affect the turnover of the GFP-htt-28Q protein.
These results provide mechanistic insight into our previous findings where we showed that overexpression of ubiquilin reduces protein aggregates and toxicity of expanded polyglutamine proteins [15]. They suggest that the simplest explanation for its protective role is that ubiquilin possesses properties that enable it not only to bind selectively to expanded polyglutamine proteins but also to target the expanded proteins for degradation.
We believe that the UBA and UBL domains of ubiquilin are likely to play fundamental roles in the selective recognition and disposal of expanded polyglutamine proteins. We propose the UBA domain of ubiquilin, which is a domain know to bind ubiquitin moieties, most likely binds the expanded polyglutamine proteins, via interaction with the polyubiquitin chain conjugated on the proteins. Proteins with longer polyglutamine tracts are more likely to be ubiquitinated than proteins with shorter polyubiquitin tracts, because of the increased propensity of expanded polyglutamine proteins to misfold and aggregate, causing them to be targeted for ubiquitination by the quality control system of the cell. Consistent with this hypothesis we found a direct correlation in the amount of ubiquilin that coimmuoprecipitated with GFP-htt polygultamine proteins and the extent of polyubiquitination of the proteins. Additionally, as we have shown here ubiquilin colocalized extensively with GFP-aggregates formed by the expanded polyglutamine 74Q proteins in cells but not with GFP-htt-28Q proteins that did not form aggregates. Moreover, in previous studies we found that ubiquilin colocalizes with ubiquitin-positive puncta in cells in accord with its ability to bind polyubiquitin species [14].
On the other hand, we propose that the UBL domain of ubiquilin most likely facilitates the delivery of the ubiquitinated proteins to the proteasome for degradation. This hypothesis is based on the ability of the UBL domain of ubiquilin to bind the 5Sa cap of the proteasome [13] [16]. In accord with this idea we showed previously that ubiquilin overexpression leads to a reduction in both the number and amount of polyglutamine aggregates in cells caused by expanded polyglutamine proteins, whereas a reduction of ubiquilin level by RNA interference produced the opposite effects [15].
The conclusion derived from the results shown here provides strong evidence that methods to elevate ubiquilin levels in cells could aid in the selective elimination of proteins containing expanded potentially toxic and misfolded polyglutamine tracts, without affecting proteins containing a normal range of polyglutamine repeats. The ability of ubiquilin to differentially discriminate between expanded toxic and normal non-toxic polyglutamine proteins could be very useful for treating polyglutamine expansion disorders which are generally linked to mutation, or inheritance, of only one mutant copy of an expanded CAG allele. In such heterozygotes, an elevation of ubiquilin levels could potentially eliminate only the proteins encoded by the expanded CAG allele, without affecting loss and the beneficial affects of the proteins encoded by the non-expanded CAG allele. This protective effect may not be restricted to treatment of only polyglutamine disorders as we have found that ubiquilin can protect cells against toxicity caused by other types of misfolded proteins, such as polyalanine expansion [18]. In summary, our results shown here suggest that elevating ubiquilin levels could provide a useful strategy to selectively remove misfolded ubiquitinated proteins that are thought build-up in cells and cause several human diseases.
Acknowledgments
This work was supported by an NIH grant GM066287 to MJM.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. The Huntington's Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 2.Bates G. Huntingtin aggregation and toxicity in Huntington's disease. Lancet. 2003;361:1642–1644. doi: 10.1016/S0140-6736(03)13304-1. [DOI] [PubMed] [Google Scholar]
- 3.Wexler NS, Lorimer J, Porter J, Gomez F, Moskowitz C, Shackell E, Marder K, Penchaszadeh G, Roberts SA, Gayan J, Brocklebank D, Cherny SS, Cardon LR, Gray J, Dlouhy SR, Wiktorski S, Hodes ME, Conneally PM, Penney JB, Gusella J, Cha JH, Irizarry M, Rosas D, Hersch S, Hollingsworth Z, MacDonald M, Young AB, Andresen JM, Housman DE, De Young MM, Bonilla E, Stillings T, Negrette A, Snodgrass SR, Martinez-Jaurrieta MD, Ramos-Arroyo MA, Bickham J, Ramos JS, Marshall F, Shoulson I, Rey GJ, Feigin A, Arnheim N, Acevedo-Cruz A, Acosta L, Alvir J, Fischbeck K, Thompson LM, Young A, Dure L, O'Brien CJ, Paulsen J, Brickman A, Krch D, Peery S, Hogarth P, Higgins DS, Jr., Landwehrmeyer B. Venezuelan kindreds reveal that genetic and environmental factors modulate Huntington's disease age of onset. Proc Natl Acad Sci U S A. 2004;101:3498–3503. doi: 10.1073/pnas.0308679101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riley BE, Orr HT. Polyglutamine neurodegenerative diseases and regulation of transcription: assembling the puzzle. Genes Dev. 2006;20:2183–2192. doi: 10.1101/gad.1436506. [DOI] [PubMed] [Google Scholar]
- 5.Bates G, Harper P, Jones L. Huntington's Disease. 3rd Ed ed. Oxford University Press; Oxford: 2002. [Google Scholar]
- 6.Michalik A, Van Broeckhoven C. Pathogenesis of polyglutamine disorders: aggregation revisited. Hum Mol Genet. 2003;12(Spec No 2):R173–186. doi: 10.1093/hmg/ddg295. [DOI] [PubMed] [Google Scholar]
- 7.Doi H, Mitsui K, Kurosawa M, Machida Y, Kuroiwa Y, Nukina N. Identification of ubiquitin-interacting proteins in purified polyglutamine aggregates. FEBS Lett. 2004;571:171–176. doi: 10.1016/j.febslet.2004.06.077. [DOI] [PubMed] [Google Scholar]
- 8.Mah AL, Perry G, Smith MA, Monteiro MJ. Identification of ubiquilin, a novel presenilin interactor that increases presenilin protein accumulation. J Cell Biol. 2000;151:847–862. doi: 10.1083/jcb.151.4.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elsasser S, Finley D. Delivery of ubiquitinated substrates to protein-unfolding machines. Nat Cell Biol. 2005;7:742–749. doi: 10.1038/ncb0805-742. [DOI] [PubMed] [Google Scholar]
- 10.Wu AL, Wang J, Zheleznyak A, Brown EJ. Ubiquitin-related proteins regulate interaction of vimentin intermediate filaments with the plasma membrane. Mol Cell. 1999;4:619–625. doi: 10.1016/s1097-2765(00)80212-9. [DOI] [PubMed] [Google Scholar]
- 11.Conklin D, Holderman S, Whitmore TE, Maurer M, Feldhaus AL. Molecular cloning, chromosome mapping and characterization of UBQLN3 a testis-specific gene that contains an ubiquitin-like domain. Gene. 2000;249:91–98. doi: 10.1016/s0378-1119(00)00122-0. [DOI] [PubMed] [Google Scholar]
- 12.Kleijnen MF, Shih AH, Zhou P, Kumar S, Soccio RE, Kedersha NL, Gill G, Howley PM. The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol Cell. 2000;6:409–419. doi: 10.1016/s1097-2765(00)00040-x. [DOI] [PubMed] [Google Scholar]
- 13.Kleijnen MF, Alarcon RM, Howley PM. The ubiquitin-associated domain of hPLIC-2 interacts with the proteasome. Mol Biol Cell. 2003;14:3868–3875. doi: 10.1091/mbc.E02-11-0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Massey LK, Mah AL, Ford DL, Miller J, Liang J, Doong H, Monteiro MJ. Overexpression of ubiquilin decreases ubiquitination and degradation of presenilin proteins. J Alzheimers Dis. 2004;6:79–92. doi: 10.3233/jad-2004-6109. [DOI] [PubMed] [Google Scholar]
- 15.Wang H, Lim PJ, Yin C, Rieckher M, Vogel BE, Monteiro MJ. Suppression of polyglutamine-induced toxicity in cell and animal models of Huntington's disease by ubiquilin. Hum Mol Genet. 2006;15:1025–1041. doi: 10.1093/hmg/ddl017. [DOI] [PubMed] [Google Scholar]
- 16.Ko HS, Uehara T, Tsuruma K, Nomura Y. Ubiquilin interacts with ubiquitylated proteins and proteasome through its ubiquitin-associated and ubiquitin-like domains. FEBS Lett. 2004;566:110–114. doi: 10.1016/j.febslet.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 17.Raasi S, Varadan R, Fushman D, Pickart CM. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat Struct Mol Biol. 2005;12:708–714. doi: 10.1038/nsmb962. [DOI] [PubMed] [Google Scholar]
- 18.Wang H, Monteiro MJ. Ubiquilin overexpression reduces GFP-polyalanine-induced protein aggregates and toxicity. Exp Cell Res. 2007 doi: 10.1016/j.yexcr.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]