Abstract
The bacteriophage T4 capsid contains two accessory surface proteins, the small outer capsid protein (Soc, 810 copies) and the highly antigenic outer capsid protein (Hoc, 155 copies). As these are dispensable for capsid formation, they can be used for displaying proteins and macromolecular complexes on the T4 capsid surface. Anthrax toxin components were attached to the T4 capsid as a fusion protein of the N-terminal domain of the anthrax lethal factor (LFn) with Soc. The LFn-Soc fusion protein was complexed in vitro with Hoc−Soc− T4 phage. Subsequently, anthrax protective antigen, PA63, heptamers were attached to the exposed LFn domains. A cryo-electron microscopy study of the decorated T4 particles shows the complex of PA63 heptamers with LFn-Soc on the phage surface. Although the cryo-electron microscopy reconstruction is unable to differentiate on its own between different proposed models of the anthrax toxin, the density is consistent with a model that had predicted the orientation and position of three LF molecules bound to one PA63 heptamer.
Keywords: Anthrax, bacteriophage T4, cryo-EM, phage display, vaccine
Introduction
Bacteriophage T4 is a large, double-stranded DNA virus that belongs to the Myoviridae family and infects Escherichia coli. The T4 virion consists of a capsid and a contractile tail terminating in a baseplate to which is attached six long tail fibers (Eiserling and Black, 1994). The T4 head is a prolate icosahedron (Fig. 1A) with a length of 1200 Å and a width of 860 Å (Fokine et al., 2004). The major capsid protein, gp23* (48.7 kDa, 930 copies per capsid), forms a hexagonal lattice with triangulation numbers (Caspar and Klug, 1962) Tend = 13 (laevo) in the icosahedral caps and Tmid = 20 in the cylindrical midsection (Fokine et al., 2004). The distance between adjacent gp23* hexameric capsomers is ∼140 Å. Eleven vertices of the capsid are occupied by pentamers of the special vertex protein gp24* (47.2 kDa), whereas the 12th vertex, occupied by a gp20 dodecamer, is the portal for DNA entry, tail attachment, and DNA exit (Driedonks et al., 1981). Gp24* and gp23* have a protein fold similar to that of the bacteriophage HK97 capsid protein (Fokine et al., 2005).
Figure 1.

Cryo-EM reconstructions. (A) The wild-type T4 capsid at 22 Å resolution (Figure from reference 2). (B) The T4 capsid with attached (PA63)7 and LFnSoc molecules at 35 Å resolution. The gp23* hexamers are shown in blue, the gp24* pentamers in magenta, the Hoc molecules in yellow, and the Soc molecules in white. Protrusions corresponding to the anthrax toxin proteins are colored in pink.
A unique feature of T4 architecture is the presence of two accessory proteins, the highly antigenic outer capsid protein (Hoc, 39.1 kDa) and the small outer capsid protein (Soc, 9.7 kDa), which decorate the outside of the capsid shell (Figs. 1A and 2A) (Fokine et al., 2004; Ishii and Yanagida, 1977; Olson et al., 2001). Both Hoc and Soc are dispensable for capsid formation and able to bind to the capsid during the final stages of capsid maturation. Soc helps to stabilize the capsid against extremes of pH and temperature, whereas Hoc has only a marginal effect on capsid stability (Steven et al., 1992). One Hoc molecule binds at the center of each gp23* hexamer, giving 155 Hoc molecules per T4 capsid. The rod-like Soc molecules form a nearly continuous mesh on the capsid surface that encircles the gp23* hexamers (Figs. 1A and 2A). The Soc molecules bind between gp23* hexamers, but not between gp23* hexamers and gp24* pentamers or between gp23* hexamers and the gp20 dodecameric portal protein. The total number of Soc molecules on the T4 capsid is 810.
Figure 2.
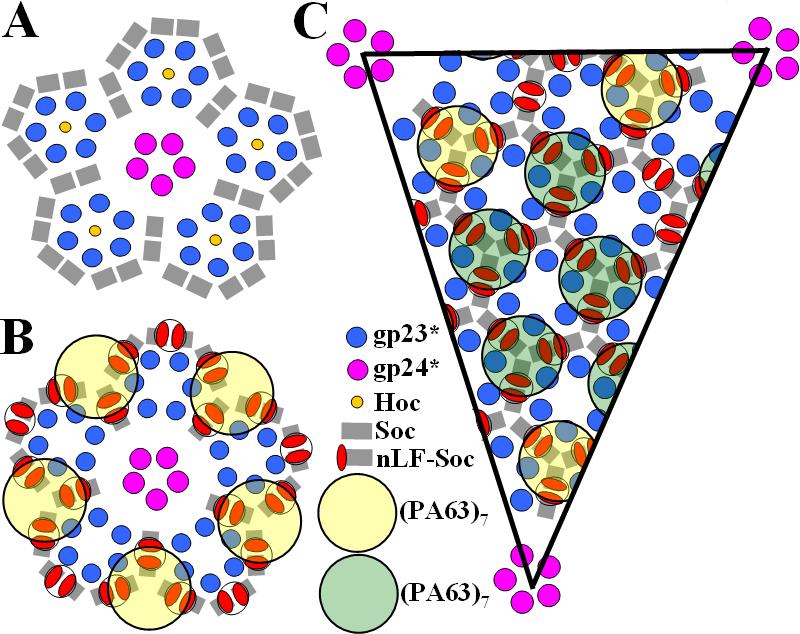
Schematic representation of the distribution of proteins on the T4 capsid surface. (A) View down a 5-fold axis of a wild-type virus and (B) showing also the attached (PA63)7-(LFn-Soc)3 complexes. (C) One possible distribution of the (PA63)7-(LFn-Soc)3 complexes in the equatorial region of the T4 capsid. The yellow rings show the higher occupancy positions of (PA63)7 in the vicinity of the 5-fold vertices, whereas the green rings show a possible distribution of the less occupied sites.
Bacillus anthracis, the causative agent of anthrax, secretes three proteins, namely protective antigen (PA), lethal factor (LF), and edema factor (EF), that assemble into toxic complexes on mammalian cell surfaces. The assembly of these complexes starts with binding PA to a cellular receptor (Bradley et al., 2001), followed by cleavage of PA by furin (a membrane-bound protease) causing removal of a 20 kDa N-terminal PA fragment. The remaining receptor-bound 63 kDa fragment of PA (PA63) oligomerizes to form a ring-shaped heptamer (Milne et al., 1994). The PA63 heptamer, (PA63)7, then interacts with the N-terminal domain of LF (LFn) and/or EF (EFn) to form the biologically active anthrax toxin complex (Cunningham et al., 2002; Leppla, 2006; Melnyk and Collier, 2006). The assembled toxin can enter the cell via receptor-mediated endocytosis. The intracellular delivery of LF, a Zn-metallo protease, and EF, an adenylate cyclase, dramatically alters the cellular metabolism, eventually causing cell death (Leppla, 2006).
Currently available information on the stoichiometry of the anthrax toxin complex is contradictory. Originally, Singh et al. (Singh et al., 1999) showed by non-denaturing gel electrophoresis that each PA63 subunit in the heptamer can bind one LF molecule. Mogridge et al.(Mogridge, Cunningham, and Collier, 2002) studied the stoichiometry of the anthrax toxin by measurements of isotope ratios in complexes assembled from differently labeled toxin subunits. Also, they determined the molecular weights of the complexes using multi-angle laser light scattering. Both approaches yielded the value of three molecules of LF or EF per PA63 heptamer. However, cryo-electron microscopy (cryo-EM) reconstruction of the complex of PA63 heptamer with LF showed only one LF molecule (Ren et al., 2004; Tama et al., 2006). Recent studies of the interactions between PA63 and LF using peptide amide hydrogen/deuterium exchange mass spectrometry and directed mutagenesis (Melnyk et al., 2006) supported the finding that PA63 heptamer can bind a maximum of three molecules of LF or EF (Mogridge, Cunningham, and Collier, 2002). The previously determined crystal structures of PA63 heptamers (Lacy et al., 2004) and LF (Pannifer et al., 2001) were used to construct a model of the (PA63)7-(LF)3 complex, consistent with mutational studies (Lacy et al., 2005).
In this paper, we report a cryo-EM study of T4 particles associated with PA63 heptamers complexed with LFn-Soc. A fusion protein between LFn (residues 1 to 264) and full-length Soc (70 residues) was constructed, with a hexa-histidine N-terminal tag and a four-residue poly-Ala linker. The LFn-Soc molecules were attached in vitro to a Hoc−Soc− T4 phage mutant. The anthrax PA63 heptamers were then attached to the capsid-exposed LFn domains. This represents probably the first structure of a phage-displayed macromolecular complex and favors the (PA63)7-(LF)3 model of the anthrax toxin (Mogridge, Cunningham, and Collier, 2002; Singh et al., 1999).
Results and discussion
The cryo-EM image of the T4 phage, with attached LFn-Soc and (PA63)7 molecules (Fig. 3), shows a pattern of roughly ring-shaped protrusions on the virus capsid. The height of the protrusions measured from the capsid surface is ∼120 Å and their width is ∼120 Å. The size and the shape of the protrusions are approximately consistent with the diameter model of (PA63)7-(LFn)3 complex (Lacy et al., 2005).
Figure 3.
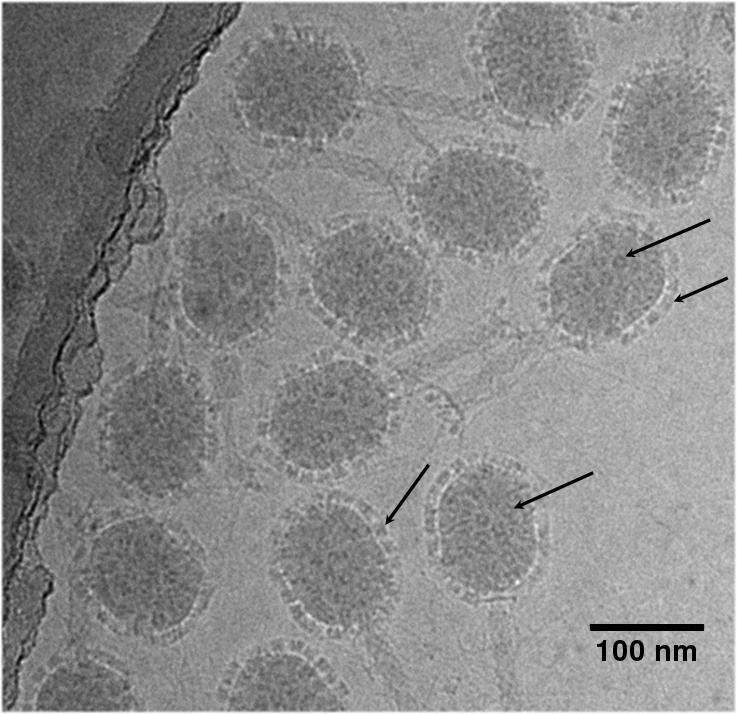
Cryo-EM image of T4 phage decorated with anthrax proteins. Note the position of the PA63 heptamers covering all parts of the capsid (see arrows).
The three-dimensional reconstruction of the T4 phage decorated with the anthrax proteins (Fig. 1B) has 35 Å resolution and shows two types of protrusions on the viral capsid. However, there is no significant structural change in the gp23*/gp24* protein shell compared to the wild-type T4 capsid (Fokine et al., 2004) (Fig. 1A). Consistent with expectations on using a Hoc−Soc− T4 phage, the reconstruction shows no evidence for the presence of Hoc molecules in the centers of the gp23* hexamers.
The smaller protrusions are located at the dimer interfaces between adjacent Soc molecules, mostly in the equatorial region of the capsid, with one protrusion per two Soc molecules (Figs. 1B and 2B). As LFn is fused to the N terminus of Soc, the N termini of the Soc molecules must be situated close to the Soc dimer interfaces on the T4 capsid. Hence, each protrusion at the Soc dimer interfaces should represent two LFn molecules. This is consistent with biochemical data (Li et al., 2006), which show that there are about 760 out of the possible 810 Soc molecule binding sites that should be occupied by the LFn-Soc fusion protein. However, the dimensions of these protrusions are too small to accommodate two LFn molecules, indicating that the LFn-Soc structure might be flexible at the poly-Ala linker region between the LFn and Soc domains. On the other hand, the height of the protrusions measured from the surface of the gp23* capsid shell is ∼75 Å, which is longer than the dimensions of the LFn molecule (∼50 Å). This suggests the density of these protrusions has a contribution from PA63 molecules.
The larger, ring-shaped protrusions in the cryo-EM reconstruction are visible only in the vicinity of the 5-fold vertices (Figs. 1B and 2B). These protrusions are centered on the quasi-3-fold axes between three adjacent gp23* hexamers. In contrast to the cryoEM reconstruction, the (PA63)7 rings are visible on all parts of the T4 capsid surface in the unprocessed EM micrograph images (Fig. 3). The apparent absence of PA63 heptamers in the reconstruction (other than in the immediate vicinity around the 5-fold vertices), as opposed to their presence in the unprocessed EM images, must be the result of averaging between occupied and unoccupied sites in combining the images to produce the three-dimensional reconstruction. The remaining five protrusions in the vicinity of each 5-fold vertex are close to the Soc trimeric binding sites located around the 5-fold vertices.
The simplest interpretation would be that the observed protrusions are the PA63 heptamers, each bound to three LFn-Soc molecules. Indeed, the predicted model (Lacy et al., 2005; Melnyk et al., 2006) of the anthrax toxin is consistent with the size of each of the observed protrusions around the 5-fold vertices. According to this interpretation the upper part of the large protrusion can be attributed to the PA63 heptamer (Figs. 1B, 2B, and 4). Each protrusion has three LFn ‘legs’ associated with the capsid surface (Figs. 2B and 4) close to the dimer interface of Soc molecules (Fig. 4). The density of each leg presumably has contributions from two LFn molecules. However, as the density corresponding to the LFn legs is not large enough to accommodate two LFn molecules, presumably only one of the LFn molecules in each leg is complexed with (PA63)7, the other LFn molecule being disoriented. Another possible interpretation is that each of the large protrusions represents the averaged density from several PA63 heptamers bound randomly to different LFn molecules. To support the first of the two proposed interpretations, the density of the three-dimensional reconstruction was projected onto a few randomly-selected EM images, given their orientation with respect to the reconstructed image. In most cases the PA63 heptamers observable in cryo-EM images superimpose onto possible LFn-Soc trimeric binding sites.
Figure 4.

Stereodiagrams showing the cryoEM density corresponding to the anthrax toxin complex. The Cα backbone (Lacy et al., 2005; Melnyk et al., 2006) is superimposed on the density with (PA63)7 in yellow and the three LFn molecules in white. (A) Side view showing also the site of attachment on the viral surface, (B) view from inside the virus showing the three (LFn) molecules
The symmetry mismatch between the 7-fold PA63 heptamer and the quasi-3-fold attachment site by three LFn “legs” suggests that there could be three possible orientations for binding each heptamer. Therefore, the result of averaging different particles is likely to generate protrusions that are essentially 3-fold averaged. This might explain why the large protrusions do not have visible 7-fold symmetry in the reconstruction. Thus, it is not possible to determine from the cryoEM reconstruction alone whether the anthrax PA63 heptamer is bound to one (Ren et al., 2004; Tama et al., 2006) or to three (Cunningham et al., 2002; Lacy et al., 2005; Melnyk and Collier, 2006) LFn molecules. Nevertheless, the atomic model (Lacy et al., 2005) of (PA63)7–(LFn)3 is roughly consistent with the density of the protrusions bound to three LFn legs (Fig. 4) in showing the relative orientation of the LFn molecules relative to the PA63 ring complex. The agreement between the two independent approaches, cryoEM image reconstruction and structural modeling by Lacy et al. (2005), suggests that the (PA63)7–(LFn)3 anthrax toxin stoichiometry is likely to be correct.
The distance between the centers of adjacent Soc trimers (corresponding to the center of PA63 heptamers) is ∼80 Å, whereas the diameter of a (PA63)7 ring is ∼120 Å. Hence, the large diameter of the PA63 heptamer rings results in steric conflict between neighboring triplets of LFn sites (Fig. 2C). Presumably, in different phage particles, the PA63 heptamers bind randomly to different LFn molecules at alternative trimeric Soc binding sites. Averaging between particles during the reconstruction procedure would result in partial PA63 heptamer occupancy at the various sites (Harris et al., 2006). Furthermore, the complex superpositions would average over three possible LFn molecules that might be binding (PA63)7, accounting for the irregular appearance of the heptamers. The steric conflicts would also account for the absence of significant (PA63)7 density at sites other than in the vicinity of the 5-fold vertices in the EM reconstruction (Figs. 1B, 2B, and 2C) (see above). The steric conflicts imply that the average number of attached PA63 heptamers per phage particle would be approximately 100, smaller than the number of possible Soc binding triplets (250), resulting in the higher occupancy of sites around these vertices. In contrast, steric hindrance between neighboring (PA63)7 rings will be greatest in the equatorial region of a prolate capsid, resulting in the lowest occupancies of the heptamer rings.
A computation, assuming equal probability of binding to trimeric Soc sites, showed that, as a result of steric hindrance, the average occupancies of these sites by PA63 heptamers would be about 0.4 in general positions and about 0.5 at the five closest sites around the 5-fold vertices. However, the cryo-EM reconstruction shows greater differentiation of occupancy between the two types of sites. This indicates that there are other factors in addition to steric hindrance that control the degree of occupancy. For instance, the difference in capsid curvature close to the 5-fold vertices leads to different angular relationship of the three LFn molecules. This might be better for heptamer binding to three LFn molecules, or, if only one LFn molecule binds to each heptamer for avoiding steric hinderance between unbound LFn molcules and bound PA63 heptamers.
This study suggests a potential use of T4 phage display for structural studies of protein complexes attached to the surface of a virus. More significantly, this work represents the first cryo-EM reconstruction of a phage display system. The reconstruction shows the presence of the displayed antigens on the T4 capsid surface with a high copy number. The T4 system is a stable phage platform for a high-density display of large, oligomeric protein complexes and could provide potential applications for vaccine development.
Materials and methods
Preparation of the Hoc−Soc− T4 particles with attached (PA63)7 and LFn-Soc was performed as described by Li et al (Li et al., 2006). About 5 × 1010 purified Hoc−SocHoc− phage particles were centrifuged at 16,000g for 40 min. The pellets were resuspended in assembly buffer (50 mM sodium phosphate pH 7.0, 75 mM NaCl, and 1 mM MgSO4) and incubated with 150 μg of purified LFn-Soc fusion protein in a total reaction mixture of 1 ml at 37°C for 45 min (construction and characterization of LFn-Soc was described by Li et al (Li et al., 2006)). The LFn–Soc–T4 phage was sedimented by centrifugation and was washed twice with excess binding buffer (20 mM Tris-Cl pH 9.0, 100 mM NaCl, and 2 mg/ml CHAPS) to remove any unbound or loosely trapped protein. The phage pellet was resuspended in 500 μl of the same buffer, and PA63 heptamers were then assembled by incubating LFn–Soc–T4 with 500 μg of PA63 at 37°C for 45 min, which is equivalent to about 20:1 excess of (PA63) 7 heptamers to LFn-Soc molecules exposed on the phage surface. The resultant phage particles were sedimented and washed as above, and the phage pellet was resuspended in 20 μl of binding buffer.
Low-dose cryo-EM was performed as described by Baker et al (Baker, Olson, and Fuller, 1999). Images of the frozen-hydrated sample were recorded on Kodak film using a Philips (Eindhoven, The Netherlands) CM300 FEG microscope at a magnification of 47,000 and an irradiation dose of ∼20 electrons per Å2 using a defocus distance of 1-3 μm. The micrographs were digitized with a ZI scanner and used to select 722 phage particles with the help of the program ROBEM (http://bilbo.bio.purdue.edu/~workshop/help_robem).
The three-dimensional image reconstructions were made with the program SPIDER (Frank et al., 1996) using the projection-matching technique to determine the orientation of the imaged particles. The reconstruction of the native bacteriophage T4 capsid (Fokine et al., 2004), represented on a 6 Å grid, was used as the initial model. Thirty cycles of image reconstruction were performed imposing D5 symmetry averaging. The final resolution of the reconstruction was 35 Å using a Fourier shell correlation cutoff of 0.5.
The atomic coordinates of the PA63 heptamers and LF were taken from the Protein Data Bank (Berman et al., 2000) (PDB accession numbers 1TZO and 1J7N) to model the structure of the (PA63)7-(LFn)3 complex. The structure of LFn (residues 27-264) was docked manually onto the structure of PA63 heptamers.
Data deposition
The cryo-EM map of the T4 head complexed with the anthrax proteins has been deposited in the Electron Microscopy Data Bank (accession number EMD-1280).
Acknowledgments
The authors thank Dr. Sathish Shivachandra (CUA) for discussions and assistance and Dr. Stephan Leppla (NIH) for reagents and discussions. We are grateful to Sheryl Kelly, Cheryl Towell, and Sharon Wilder for the preparation of the manuscript. This work was supported by an NSF grant (MCB-0443899) and an HFSP grant (RGP-28/200) to MGR and an NIH grant (AI056443) to VBR. In addition, the work benefited from a W. M. Keck Foundation grant (981675) to MGR for the purchase of a CM300 FEG electron microscope.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baker TS, Olson NH, Fuller SD. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiol. Mol. Biol. Rev. 1999;63:862–922. doi: 10.1128/mmbr.63.4.862-922.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucl. Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley KA, Mogridge J, Mourez M, Collier RJ, Young JA. Identification of the cellular receptor for anthrax toxin. Nature (London) 2001;414:225–229. doi: 10.1038/n35101999. [DOI] [PubMed] [Google Scholar]
- Caspar DLD, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harbor Symp. Quant. Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- Cunningham K, Lacy DB, Mogridge J, Collier RJ. Mapping the lethal factor and edema factor binding sites on oligomeric anthrax. Proc. Natl. Acad. Sci. U.S.A. 2002;99:7049–7053. doi: 10.1073/pnas.062160399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driedonks RA, Engel A, ten Heggeler B, van Driel R. Gene 20 product of bacteriophage T4. Its purification and structure. J. Mol. Biol. 1981;152:641–662. doi: 10.1016/0022-2836(81)90121-2. [DOI] [PubMed] [Google Scholar]
- Eiserling FA, Black LW. Pathways in T4 morphogenesis. In: Karam JD, editor. Molecular Biology of Bacteriophage T4. American Society for Microbiology; Washington, D.C.: 1994. pp. 209–212. [Google Scholar]
- Fokine A, Chipman PR, Leiman PG, Mesyanzhinov VV, Rao VB, Rossmann MG. Molecular architecture of the prolate head of bacteriophage T4. Proc. Natl. Acad. Sci. U.S.A. 2004;101:6003–6008. doi: 10.1073/pnas.0400444101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokine A, Leiman PG, Shneider MM, Ahvazi B, Boeshans KM, Steven AC, Black LW, Mesyanzhinov VV, Rossmann MG. Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proc. Natl. Acad. Sci. U.S.A. 2005;102:7163–7168. doi: 10.1073/pnas.0502164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Harris A, Belnap DM, Watts NR, Conway JF, Cheng N, Stahl SJ, Vethanayagam JG, Wingfield PT, Steven AC. Epitope diversity of hepatitis B virus capsids: quasi-equivalent variations in spike epitopes and binding of different antibodies to the same epitope. J. Mol. Biol. 2006;355(3):562–576. doi: 10.1016/j.jmb.2005.10.035. [DOI] [PubMed] [Google Scholar]
- Ishii T, Yanagida M. The two dispensable structural proteins (soc and hoc) of the T4 phage capsid: their properties, isolation and characterization of defective mutants, and their binding with the defective heads in vitro. J. Mol. Biol. 1977;109:487–514. doi: 10.1016/s0022-2836(77)80088-0. [DOI] [PubMed] [Google Scholar]
- Lacy DB, Lin HC, Melnyk RA, Schueler-Furman O, Reither L, Cunningham K, Baker D, Collier RJ. A model of anthrax toxin lethal factor bound to protective antigen. Proc. Natl. Acad. Sci. U.S.A. 2005;102:16409–16414. doi: 10.1073/pnas.0508259102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacy DB, Wigelsworth DJ, Melnyk RA, Harrison SC, Collier RJ. Structure of heptameric protective antigen bound to an anthrax toxin receptor: a role for receptor in pH-dependent pore formation. Proc. Natl. Acad. Sci. U.S.A. 2004;101:13147–13151. doi: 10.1073/pnas.0405405101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppla SH. Bacillus anthracis toxins. In: Alouf JE, Popoff MR, editors. The Comprehensive Sourcebook of Bacterial Protein Toxins. Academic Press; Burlington, MA: 2006. pp. 323–347. [Google Scholar]
- Li Q, Shivachandra SB, Leppla SH, Rao VB. Bacteriophage T4 capsid: a unique platform for efficient surface assembly of macromolecular complexes. J. Mol. Biol. 2006;363:577–588. doi: 10.1016/j.jmb.2006.08.049. [DOI] [PubMed] [Google Scholar]
- Melnyk RA, Collier RJ. A loop network within the anthrax toxin pore positions the phenylalanine clamp in an active conformation. Proc. Natl. Acad. Sci. U.S.A. 2006;103:9802–9807. doi: 10.1073/pnas.0604000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnyk RA, Hewitt KM, Lacy DB, Lin HC, Gessner CR, Li S, Woods VL, Jr., Collier RJ. Structural determinants for the binding of anthrax lethal factor to oligomeric protective antigen. J. Biol. Chem. 2006;281:1630–1635. doi: 10.1074/jbc.M511164200. [DOI] [PubMed] [Google Scholar]
- Milne JC, Furlong D, Hanna PC, Wall JS, Collier RJ. Anthrax protective antigen forms oligomers during intoxication of mammalian cells. J. Biol. Chem. 1994;269:20607–20612. [PubMed] [Google Scholar]
- Mogridge J, Cunningham K, Collier RJ. The lethal and edema factors of anthrax toxin bind only to oligomeric forms of the protective antigen. Proc. Natl. Acad. Sci. U.S.A. 2002;99:7045–7048. doi: 10.1073/pnas.052160199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson NH, Gingery M, Eiserling FA, Baker TS. The structure of isometric capsids of bacteriophage T4. Virology. 2001;279:385–391. doi: 10.1006/viro.2000.0735. [DOI] [PubMed] [Google Scholar]
- Pannifer AD, Wong TY, Schwarzenbacher R, Renatus M, Petosa C, Bienkowska J, Lacy DB, Collier RJ, Park S, Leppla SH, Hanna PC, Liddington RC. Crystal structure of the anthrax lethal factor. Nature (London) 2001;414:229–233. doi: 10.1038/n35101998. [DOI] [PubMed] [Google Scholar]
- Ren G, Quispe J, Leppla SH, Mitra AK. Large-scale structural changes accompany binding of lethal factor to anthrax protective antigen: a cryo-electron microscopic study. Structure. 2004;12:2059–2066. doi: 10.1016/j.str.2004.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh Y, Klimpel KR, Goel S, Swain PK, Leppla SH. Oligomerization of anthrax toxin protective antigen and binding of lethal factor during enodcytic uptake into mammalian cells. Infect. Immun. 1999;67:1853–1859. doi: 10.1128/iai.67.4.1853-1859.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steven AC, Greenstone HL, Booy FP, Black LW, Ross PD. Conformational changes of a viral capsid protein. Thermodynamic rationale for proteolytic regulation of bacteriophage T4 capsid expansion, cooperativity, and super-stabilization by soc binding. J. Mol. Biol. 1992;228:870–884. doi: 10.1016/0022-2836(92)90871-g. [DOI] [PubMed] [Google Scholar]
- Tama F, Ren G, Brooks CL, III, Mitra AK. Model of the toxic complex of anthrax: responsive conformational changes in both the lethal factor and the protective antigen heptamer. Prot. Sci. 2006;15:2190–2200. doi: 10.1110/ps.062293906. [DOI] [PMC free article] [PubMed] [Google Scholar]


