Abstract
Accurate protein identification sometimes requires careful discrimination between closely related protein isoforms that may differ by as little as a single amino acid substitution or post-translational modification. The ABRF Proteomics Research Group sent a mixture of three picomoles each of three closely related proteins to laboratories who requested it in the form of intact proteins, and participating laboratories were asked to identify the proteins and report their results. The primary goal of the ABRF-PRG04 Study was to give participating laboratories a chance to evaluate their capabilities and practices with regards to sample fractionation (1D- or 2D-PAGE, HPLC, or none), protein digestion methods (in-solution, in-gel, enzyme choice), and approaches to protein identification (instrumentation, use of software, and/or manual techniques to facilitate interpretation), as well as determination of amino acid or post-translational modifications. Of the 42 laboratories that responded, 8 (19%) correctly identified all three isoforms and N-terminal acetylation of each, 16 (38%) labs correctly identified two isoforms, 9 (21%) correctly identified two isoforms but also made at least one incorrect identification, and 9 (21%) made no correct protein identifications. All but one lab used mass spectrometry, and data submitted enabled a comparison of strategies and methods used.
Keywords: proteomics, protein isoforms, mass spectrometry, protein identification, post-translational modifications
A common task for proteomic core facilities, aside from the usual identification of proteins, is to locate differences between proteins of interest. These may simply be cross-species differences or modifications associated with a vital roll, for example, in protein structure and function. The investigator may have only minimal information regarding the type or location of these differences. These considerations make the ability to locate such variations very important. To make this determination, a core facility must investigate the protein in depth to obtain maximum sequence coverage, going beyond a cursory matching of the protein to an entry in a sequence database. This study was designed help evaluate the abilities of core facilities to identify closely related proteins and determine where the differences exist between them. Therefore, the primary goals of this study were to give each laboratory a chance to evaluate its capabilities and practices with regards to protein digestion methods (solution based, in gel, choice of enzymes), protein identification methods, methods for the determination of amino acid differences between protein isoforms, amino acid sequence coverage of the identified proteins, characterization of simple post-translational modifications, as well as to obtain data that would allow a comparison of the strategies used and aid in optimization of these techniques.
The Proteomics Research Group (PRG) provided laboratories that requested samples with a mixture of three intact proteins: two bovine carbonic anhydrase isoforms differing by a single amino acid substitution plus a human carbonic anhydrase. Because the sequences of these proteins can be found in the public databases, we required that the participants supply proof of the differences ascertained by tandem mass spectrometry or other means they may have used. This study related to previous PRG studies in that proteins in a mixture were to be identified and amino acid modifications or substitutions analyzed. However, in this study, unlike the past two ABRF-PRG studies,1,2 intact proteins, rather than a predigested mixture, were distributed. Furthermore, the analysis involved searching for differences between three very similar proteins, rather than modifications within the same protein. Also, because the participants did their own digests, this study addressed, to some extent, the issue of front-end sample preparation. We asked that participants return proof of the differences they determined, along with the completed online questionnaire detailing methods and strategies used by the participants for their determinations. As in past studies, the identities of the respondents were not known to members of the PRG.
MATERIALS AND METHODS
Sample Preparation
Three carbonic anhydrase protein isoforms (C6165, C2522, and C3640) were purchased from Sigma-Aldrich Co. (St. Louis, MO). Small amounts of each protein were weighed using a Cahn microbalance, and an appropriate amount of 1% acetic acid was added to produce a final concentration of 1 μg/μL. Specifically, 1.15 mg of C6165 was solubilized in 1.15 mL 1% acetic acid; 1.06 mg C2522 was solubilized in 1.06 mL 1% acetic acid; and 1.20 mg of C3640 was solubilized in 1.20 mL 1% acetic acid.
Three microliters of each protein solution or 1% acetic acid control were placed into amino acid analysis tubes, and each sample was prepared in triplicate. The samples were lyophilized in a vacuum centrifuge and sealed by wrapping in parafilm. Amino acid analysis indicated that the amount of protein present was approximately 60% by weight of the protein powders purchased from Sigma-Aldrich.
The molecular weight of each protein was calculated from the corresponding amino acid sequence. From these molecular weight values and based on the amino acid analysis, the number of picomoles per microliter was determined to be: C6165 (Mr = 29156.9; 34.30 pmol/μL), C2522 (Mr = 29024.6; 34.45 pmol/μL), and C3640 Mr = 28996.6; 34.49 pmol/μL). In order to prepare a 1 pmol/μL working solution, 29 μL of each stock solution was diluted to 1000 μL final volume. Similarly, to get a 5 pmol/μL working solution, 145 μL of each stock solution was diluted to 1000 μL final volume. The remainders of the stock solutions were stored at −20°C.
One-microliter aliquots of each working solution (1 and 5 pmol) were placed in individual 0.5-mL Eppendorf tubes, lyophilized in a vacuum centrifuge, and mailed to PRG members as test samples for preliminary characterization by 1D and 2D SDS-PAGE, in-gel and in-solution tryptic digestion, and tandem mass spectrometry using ion trap (ThermoFinnigan LCQ DECA XP Plus) and quadrupole time-of-flight (Micromass Q-TOF 1) mass spectrometry to confirm the identities and preparation of the proteins. Results were analyzed using SEQUEST (for ion-trap data) and Mascot (for Q-TOF data) database search engines.
Fresh stock solutions of each of the three proteins were prepared in 1% acetic acid, as were fresh working solutions at a concentration of 3 pmoles/μL, all as described above. Three-hundred samples were prepared by aliquoting 1 μL of each protein solution into 0.5-mL Eppendrof tubes containing 10 μL of 1% acetic acid. All samples were lyophilized in a vacuum centrifuge, and sealed by wrapping in parafilm. Lyophilized samples were then sent to requesting laboratories for protein identification. Our goal was to provide participants with the minimum amount of information about the samples that would still allow a reasonable chance of success, as this is similar to the situation encountered during the analysis of typical unknown samples by core labs. The following information about the samples and instructions were included in the letter that accompanied the samples:
Sample Information
ABRF-PRG04 contains 3 pmol each of three closely related intact proteins. The sample is supplied as a dried pellet, and can be dissolved in most common aqueous solutions. As with any real-life sample, there may also be contaminant(s).
Analysis
Identify the three most abundant protein isoforms, from up to two different species, in the sample.
Identify one post-translational modification common to all three isoforms, excluding those that may happen during sample handling such as oxidation and deamidation.
Provide discriminatory evidence for the specific identifications (e.g., MS/MS spectrum or amino acid sequence for a diagnostic peptide, exact protein mass, etc.) for each isoform and the modification.
Submit your results online by filling out the online form and by faxing up to four pieces of supporting data for the identifications (tandem mass spectra with key diagnostic fragment ions labeled, etc.) according to the instructions given below.
Participant Survey
The participant survey this year was done online using a Web-based questionnaire (SurveyMonkey.com) with user-selected codes to identify the data and preserve anonymity of the participants. Questions pertained to the manner in which the sample was prepared, methods for separation of intact proteins (if any), type of gel and stain (if applicable), type of digestion and enzyme (if done), what kind of solvents if HPLC was done, and the identification along with confidence level and percent coverage of the amino acid sequence of each protein identified. Other questions pertaining to perception of difficulty of the study, opinions about the amount of time spent on the study, reasons for success or failure, and general comments about the study were asked. In addition to the online survey, because the protein sequences could be found in public sequence databases, the participants were requested to fax supporting data for the isoforms discriminations (such as MS/MS spectra) to a third party to maintain the anonymity of the participants.
RESULTS AND DISCUSSION
Study Overview
Unlike the previous two ABRF-PRG studies,1,2 an intact protein mixture rather than a protein digest was distributed to the laboratories requesting the sample. One-hundred and six samples were sent to participating laboratories, and 42 labs returned data. By providing participants with a mixture of three closely related proteins, our goal was to evaluate the success of the protein identification as well as the sample preparation approaches such as protein separation and digestion. Also of interest were the methods used for locating amino acid differences and the correlation between protein coverage and successful discrimination.
Preliminary Characterization of the Test Sample by the Proteomics Research Group
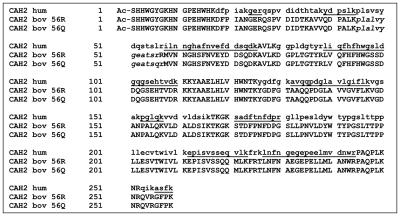
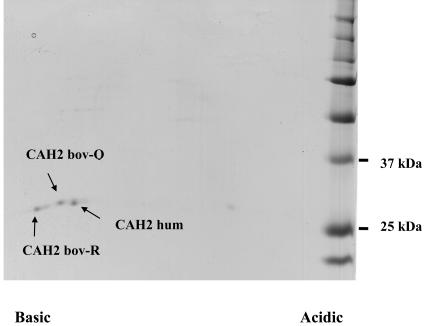
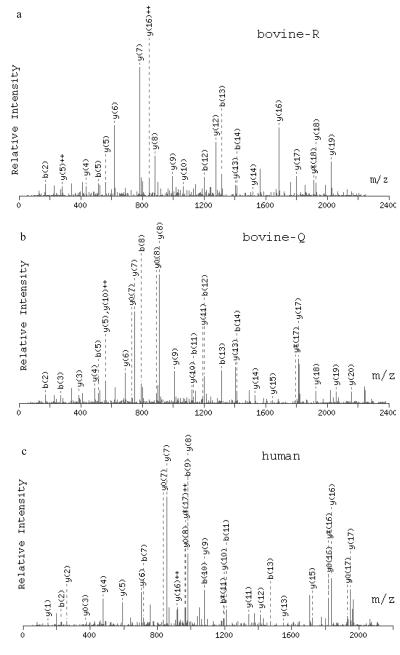
The sequences of the three carbonic anhydrase isoforms are shown in Figure 1. A Coomassie blue–stained 2D SDS-PAGE gel of 3 pmol of the protein isoforms mixture is shown in Figure 2. Representative Q-TOF MS/MS spectra for three peptides from an in-solution tryp-tic digest of the mixture are shown in Figure 3. The spectrum in Figure 3a identifies a peptide of sequence AVVQDPALKPLALVYGEATSRR that corresponds to bovine carbonic anhydrase 2 (bov-CAH2). The spectrum in Figure 3b identifies a peptide of sequence AVVQD-PALKPLALVYGEATSQR that corresponds to the same sequence from bovine CAH shown in 3a, but with an R→Q substitution at amino acid position 56 (numbering from sequence in Figure 1) that uniquely identifies the bovine bov-CAH2-Q isoform. Figure 3c shows a spectrum corresponding to a peptide of sequence ILNNGHAFN-VEFDDSQDK from human CAH2 that is not found in either of the bovine isoforms. Intact masses determined by a Research Group member were consistent with these modifications. Six study participants obtained masses for the intact protein: one by MALDI-TOF, one by ESI-TOF, and four by ESI-quadrupole-TOF. Probably because of the limited sample amount provided, most participants performed analyses at the peptide level only because of the requirement for protein identification as well as the need to provide specific information about modifications.
FIGURE 1.
Amino acid sequences of the two bovine and one human CAH2 isoforms present in the sample mixture. Tryptic peptides unique to human CAH2 are shown in small letters, and alternate peptides are underlined; peptides unique to the two bovine CAH2 isoforms are shown in small letters and italics. All isoforms were acetylated at the N-terminal serine.
FIGURE 2.
2D PAGE of 3 pmol carbonic anhydrase protein mixture. The first dimension pH gradient was 3–10 nonlinear, the second dimension SDS-PAGE gel contained 12% acrylamide. Proteins were visualized by colloidal Coomassie blue staining.
FIGURE 3.
Q-TOF MS/MS spectra of three peptides from an in-solution digest of the ABRF-PRG04 sample mixture. In each spectrum, b* or y* means b or y ion minus 17 Da; bº or yº means b or y ion minus 18 Da. (a) MS/MS spectrum of a peptide of sequence AVVQD-PALKPlAlVYGEATSRR corresponding to a tryptic peptide from bovine carbonic anhydrase 2. (b) MS/MS spectrum of a peptide of sequence AVVQDPAlK-PLALVYGEATSQR corresponding to a tryptic peptide from bovine carbonic anhydrase 2 isoform 56R→ Q (c) MS/MS spectrum of a peptide of sequence ILNNGHAFNVEFDDSQDK corresponding to a tryptic peptide from human carbonic anhydrase 2.
Sample Preparation
Results in this and following sections were obtained by study participants. A summary of sample preparation methods and protein identification results is given in Table 1. Sample solvent varied considerably, with no obvious correlation between solvent used and success in identification of the isoforms. Ammonium bicarbonate (ABC) in varying concentrations from 25 mM to 100 mM with or without additional acids or organics was the most common (18 respondents); four used sodium dodecylsulfate (SDS) solubilizing buffer; one included β-mercaptoethanol (BME); four used urea, either 8 M (most common) or 6 M; eight used trifluoroacetic acid (TFA) or formic acid (FA), again at varying concentrations, with or without other components; eight used acetonitrile (ACN) at various concentrations; and five used water alone.
TABLE 1.
Summary of Sample Preparation Methods and Protein Identification Results
| Protein 1
|
Protein 2
|
Protein 3
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ID | Solvent | Volume/μl | Separation Method | Gel or Column Type | Staining Method | Protein 1 ID | Confidence | % Sequence Coverage | Termini Observed | Protein 2 ID | Confidence | % Sequence Coverage | Termini Observed | Protein 3 ID | Confidence | % Sequence coverage | Termini Observed |
| 3 Correct protein IDs | |||||||||||||||||
| 715 | SDS SB, pH 8.5 | 10 | SDS-PAGE | 10% Tris-Gly Standard | CBB | CAH2bov-R | P | 79 | N,C, N-Ac | CAH2bov-Q | P | 79 | N,C, N-AC | CAH2hum | P | 77 | N, N-Ac |
| 98166 | SDS SB BME | 20 | SDS-PAGE | 10-20% Tris-Gly Mini | Zinc | CAH2hum | P | 66 | N, N-Ac | CAH2bov-R | P | 65 | N, N-AC | CAH2bov-Q | P | 66 | N, N-AC |
| 20702 | 1% FA | 20 | CAH2hum | P | 63 | N, N-Ac | CAH2bov-R | P | 59 | N, N-AC | CAH2bov-Q | P | 60 | N, N-AC | |||
| 27974 | water | 20 | SDS-PAGE | 12% Bis-Tris NuPAGE | SYPRO Orange | CAH2bov-R | P | 57 | N, N-Ac | CAH2bov-Q | P | 58 | N, N-AC | CAH2hum | P | 47 | N, N-AC |
| 65213 | 100 mM ABC | 60 | CAH2bov-R | P | 58 | N, N-Ac | CAH2bov-Q | P | 58 | N, N-AC | CAH2hum | P | 51 | N, N-AC | |||
| 4343 | 25 mM ABC | 20 | CAH2bov-R | P | 52 | N, N-Ac | CAH2bov-Q | P | 52 | N, N-AC | CAH2hum | P | 52 | N, N-AC | |||
| 29103 | 100 mm ABC, 0.5 m GuHCL | 100 | CAH2bov-R | P | 43 | N, N-Ac | CAH2hum | P | 40 | N, N-AC | CAH2bov-Q | P | 39 | N, N-AC | |||
| 31113 | 50 mM ABC, pH 8 | 15 | SDS-PAGE (33% of sample) | 12% Tris-Gly mini | MS Friendly Silver | CAH2hum | P | 22 | N, N-Ac | CAH2bov-R | P | 20 | N, N-AC | CAH2bov-Q | P | 20 | N, N-AC |
| 2 Correct protein IDs | |||||||||||||||||
| 11010* | water | 10 | CAH2hum | P | 55 | N, N-Ac | CAH2bov-R | P | 49 | N, N-AC | CAH2horse | T | 31 | ||||
| 1066352 | 0.1% TFA/10% ACN (9:1) | 6 | CAH2bov | P | 42 | N, N-Ac | CAH2hum | P | 45.9 | N, N-AC | CAH2bov-Q | P | 42 | N | |||
| 21562 | 0.1% FA, 0.25% N-octylglucopyranoside | 15 | 2DE (50% of sample), RP-HPLC (33% of sample) | C8 Vydac | Colloidal CBB | CAH2bov | P | 79 | N, N-Ac | CAH2hum | P | 69 | N, N-AC | ||||
| 25519 | water | 30 | CAH2bov-R | P | 73 | N, N-Ac | CAH2hum | P | 64 | N, N-AC | N-AC | ||||||
| 11111 | 8 ,M Urea, 0.2 m Tris-HCL | 20 | CAH2bov-R | P | 63 | N, N-Ac | CAH2hum | P | 64 | N, N-AC | |||||||
| 90894 | 8 M urea in 0.4 M ABC | 10 | CAH2hum | P | 62 | N, N-Ac | CAH2bov-R | P | 66 | N, N-AC | |||||||
| 22626 | 6% ACN | 5.3 | CAH2bov | P | 57 | N, N-Ac | CAH2hum | P | 50 | N, N-AC | |||||||
| 94591 | water | 10 | CAH2hum | P | 56 | N, N-Ac | CAH2bov-R | P | 55 | N, N-AC | |||||||
| 23312 | ABC | 20 | CAH2bov | P | 55 | N, N-Ac | CAH2hum | P | 46 | N, N-AC | keratin | P | |||||
| 2115 | 50 mm ABC | 30 | SDS-PAGE | 12% Tris-Gly Mini | CBB | CAHIihum | P | 70 | CAH2bov | P | 66 | N, N-AC | |||||
| 24389 | ABC | 5 | SDS-PAGE | 12% MES Mini | SYPRO | CAH2bov | P | 46 | N, N-Ac | CAH2hum | P | 41 | N, N-AC | ||||
| 32569 | 30 | CAH2hum | P | 60 | N | CAH2bov | P | 50 | N | ||||||||
| 11787 | 25 mm ABC, 10% ACN | 50 | CAH2bov-R | P | 54 | N | CAH2hum | P | 44 | ||||||||
| 73108 | 100 mm ABC | 60 | CAH2hum | P | 38 | CAH2bov | P | 30.4 | |||||||||
| 93743 | ABC | 10 | SDS-PAGE | 15% Tris-Gly Mini | Silver | CAH2hum | P | 11 | CAH2bov-R | P | 23 | ||||||
| 10567 | Water | 25 | SDS-PAGE | 4–12% NuPAGE | Silver | CAH2bov | P | 11 | CAHiihum | T | 7 | ||||||
| 2 Correct and 1 incorrect ID | |||||||||||||||||
| 24770 | 150 | RP-HPLC | C18, 75 μm x 15 cm, Picotip 20 μm | CAH2bov | P | 46 | Phos | CAH2hum | T | 20 | |||||||
| 92711 | 8 M Urea | 8 | CAH2bov | P | 54 | N-Ac | CAH2bov | P | 46 | N-AC | CAH2hum | P | 47 | N-AC | |||
| 11735 | 5% ACN, 100 mM ABC | 19 | CAH2hum | P | 42 | N | CAH2bov-R | P | 77 | N | CAH2hum | P | 16 | ||||
| 48583 | 50 mM ABC | 10 | CAH2bov | P | 66 | N, N-Ac | CAH2hum | P | 59 | N, N-AC | CAHi1hum | T | 63 | ||||
| 69186 | Tris-HCl, 6 M urea | 5 | CAH2bov | P | 47.9 | CAH2hum | P | 48 | CAH2sheep | P | 32.4 | ||||||
| 87050 | SDS SB | 60 | SDS-PAGE | 5–20% Tris-Gly Mini | SYPRO Ruby | CAH2hum | P | 62.5 | N, N-Ac | CAH2bov | P | 22.7 | CAH2sheep | P | 15.8 | ||
| 31815 | 50 mM ABC | 20 | CAH2hum | P | 38 | N, N-Ac | CAH2sheep | P | 32 | N-AC | CAH2bov-R | P | 24 | N | |||
| 21068 | o.1% TFA, 5% ACN | 20 | CAH2bov | P | 72 | C | CAH2hum | P | 22 | N | CAH2sheep | P | 17 | ||||
| 640921 | 2% ACN, 0.1% FA | 50 | CAH2sheep | P | 50 | CAH2hum | P | 40 | CAH2bov-R | P | 67 | N, N-AC | |||||
| No IDs | |||||||||||||||||
| uicmslw | 50 mM ABC | 10 | no results | ||||||||||||||
| 80053 | 40% ACN | 50 | no results | ||||||||||||||
| 69117 | 5% FA | 10 | no results | ||||||||||||||
| 106369 | no results | ||||||||||||||||
| 13791 | 50 mM ABC | 20 | no results | ||||||||||||||
| 11747 | SDS SB | 100 | SDS-PAGE | 10% Tris-Gly | CBB | no results | |||||||||||
| 13053 | 30% ACN, 0.1% TFA | 25 | SDS-PAGE | 12% Tris-Gly Mini | CBB | no results | |||||||||||
| Only Incorrect IDs | |||||||||||||||||
| 11596 | 3% TFA | 30 | calsequestrin | P | 28 | calsequestrin (dog) | P | 25 | calsequestrin (hum) | P | 4 | ||||||
| 11128 | 25 mM ABC | 10 | BSA | P | 17 | ||||||||||||
ABC: ammonium bicarbonate
ACN: acetonitrile
FA: formic acid
GuHCl: guanidinium hydrochloride
TFA: trifluoroacetic acid
CBB: coomassie Brilliant Blue
N-Ac: N-terminal acetylation observed
Phos: phosphorylation reported
SDS SB BME: SDS sample buffer plus beta mercaptoethanol
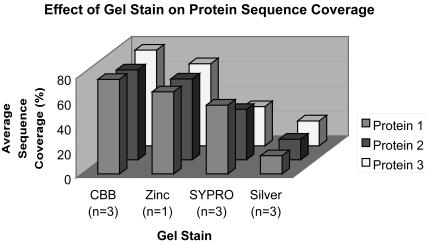
Eleven of the 42 laboratories returning data separated the proteins by 1D SDS-PAGE, and one lab used 2D SDS-PAGE. Of these twelve labs, five used Coomassie Brilliant Blue to stain the gels (two of these labs did not obtain any protein identification results), one used zinc staining, three used SYPRO orange or ruby staining, and another three used silver staining. Figure 4 shows the correlation between type of stain used and average sequence coverage obtained for each protein. This figure includes only data from the three labs that obtained results after staining with Coomassie Brilliant Blue. If data from the two labs that obtained no results after Coomassie staining (0% protein coverage) had been included, the average protein coverage for proteins 1, 2, and 3 would be 46%, 43%, and 46%, respectively, which is similar to the results obtained with Sypro staining. However, conclusions based on these data must be taken with caution, because the numbers of participants using each staining method are small and different methods, instruments, experimenters, etc., were used to identify the proteins in each experiment. For example, one of the respondents who used Coomassie blue staining used only 50% of the sample, and one who stained with silver used 33% of the sample for the SDS-PAGE separation. Of the two labs that did reverse-phase HPLC (RP-HPLC) on the intact proteins, one of the labs split the sample and did 2D electrophoresis as well. Twenty-four labs performed in-solution digestion of the unseparated mixture, while the rest either returned insufficient data to determine their methods or did not obtain any results. Prior separation of this simple mixture of three intact proteins did not correlate with superior results in correctly identifying the proteins. Half of the respondents who correctly identified all three proteins and all acetylations (four out of eight) separated the proteins before analysis, compared with 43% of labs overall.
FIGURE 4.
Effect of gel staining on sequence coverage. Average sequence coverage of identified proteins is shown for each protein, and grouped by type of stain used to visualize the proteins.
Digestion Method
Table 2 shows the protein coverage for each of the identified proteins along with the enzyme used to digest the proteins by each lab (one lab did not digest the proteins, two labs digested the proteins but reported no enzyme). Thirty laboratories used a single enzyme, while three labs used two different enzymes and three labs used three enzymes. Trypsin was the protease of choice, used exclusively by 29 labs and in combination by 5 others. Two labs used chymotrypsin, two labs used Lys-C, and three labs used Glu-C. Subtilisin, proteinase K, elastase, and Asp-N were each used one time. Sample solvent for digestions done in solution was most frequently ABC in varying concentrations from 25 mM to 100 mM, with or without additional acids or organics as noted above. Fourteen respondents used ABC; four used urea, either 8 M (most common) or 6 M; six used TFA or FA, again at varying concentrations, with or without other components; seven used ACN; and three used water. Of the eight respondents that obtained the correct identifications of the proteins along with the modifications, four did in-solution digestions. These four used the following solvents: 1% FA, 100 mM ABC, 25 mM ABC, and 100 mM ABC + 0.5 M guanidinium hydrochloride. For the four that did in-gel digestions, the following solvents were used: SDS solubilizing buffer with and without BME, water, and 50 mM ABC.
TABLE 2.
Summary of Protein Digestion Methods and Protein Sequence Coverage
| Peptide Masses Measured Without PSD or MS/MS
|
Peptide Masses Measured with PSD or MS/MS
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Protein 1
|
Protein 2
|
Protein 3
|
Protein 1
|
Protein 2
|
Protein 3
|
||||||||
| ID | Enzyme(s) used | #Pep | % Cov | #Peptides Matched | %Sequence Coverage | #Peptides Matched | %Sequence Coverage | #Peptides Matched | %Sequence Coverage | #Peptides Matched | %Sequence Coverage | #Peptides Matched | %Sequence Coverage |
| 3 Correct Protein IDs | |||||||||||||
| 715 | T | 19 | 79 | 19 | 79 | 21 | 77 | 18 | 78 | 18 | 78 | 16 | 67 |
| 98166 | T, Ch | 14 | 66 | 13 | 65 | 13 | 66 | ||||||
| 20702 | T | 15 | 63 | 15 | 59.2 | 15 | 59.6 | ||||||
| 27974 | T | 12 | 57 | 12 | 58 | 12 | 43 | 2 | 11 | 2 | 12 | 2 | 20 |
| 65213 | T, Glu-C, Ch | 11 | 58 | 11 | 58 | 12 | 51 | 6 | 28 | 6 | 28 | 8 | 30 |
| 4343 | T | 14 | 52 | 14 | 53 | 13 | 52 | ||||||
| 29103 | T | 7 | 35 | 10 | 40 | 6 | 32 | 3 | 8 | 1 | 8 | ||
| 31113 | T | 4 | 22 | 3 | 20 | 3 | 20 | ||||||
| 2 Correct Protein IDs | |||||||||||||
| 11010* | T | 8 | 30 | 7 | 34 | 0 | 0 | 6 | 28 | 5 | 20 | 4 | 15 |
| 1066352 | Lys-C | 3 | 13.9 | 0 | 0 | 18 | 42 | 13 | 45.9 | 18 | 42 | ||
| 21562 | T | 18 | 70 | 14 | 58 | 17 | 63 | 5 | 25 | ||||
| 25519 | T | 15 | 73 | 16 | 64 | ||||||||
| 11111 | T | 12 | 59 | 13 | 56 | 1 | |||||||
| 90894 | T | 18 | 61.8 | 20 | 66 | ||||||||
| 22626 | T | 10 | 56 | 12 | 50 | 5 | 27 | 8 | 30 | ||||
| 94591 | T | 9 | 53.1 | 9 | 51.9 | 1 | 2.6 | 1 | 2.6 | ||||
| 23312 | T | 12 | 55 | 8 | 46 | 4 | 23 | 6 | 25 | ||||
| 2115 | T | 16 | 70 | 14 | 68 | ||||||||
| 24389 | T, Glu-C | 10 | 45 | 11 | 41 | 10 | 45 | 11 | 41 | ||||
| 32569 | T | 10 | 50 | 5 | 28 | ||||||||
| 11787 | T | 11 | 53 | 9 | 44 | ||||||||
| 73108 | T | 10 | 44 | 9 | 30 | 6 | 38 | 5 | 30.4 | ||||
| 93743 | T | 2 | 11 | 6 | 24 | 2 | 11 | 6 | 24 | ||||
| 10567 | T | 3 | 11 | 2 | 7 | ||||||||
| 2 Correct and 1 Incorrect ID | |||||||||||||
| 24770 | Sub, ProK (pH11) | 17 | 45.8 | 2 | 7.7 | ||||||||
| 92711 | T | 8 | 54 | 6 | 46 | 8 | 47 | ||||||
| 11735 | T | 11 | 42 | 16 | 7 | 34 | 7 | 30 | 6 | 34 | |||
| 48583 | T, Glu-C, Elas | 15 | 66 | 12 | 59 | 11 | 63 | ||||||
| 69186 | T | 11 | 10 | 5 | 14 | 47.9 | 11 | 48.3 | 5 | 32.4 | |||
| 87050 | T, Lys-C, Asp-N | 12 | 62.5 | 4 | 22.7 | 4 | 15.8 | ||||||
| 31815 | T | 9 | 37 | 8 | 32 | 8 | 31 | 9 | 37 | 5 | 24 | 6 | 23 |
| 21068 | None | 30 | 73 | 10 | 22 | 5 | 17 | ||||||
| 640921 | T | 9 | 50 | 9 | 40 | 14 | 67 | 9 | 50 | 9 | 40 | 14 | 67 |
| No Identifications | |||||||||||||
| uicmslw | |||||||||||||
| 80053 | T | ||||||||||||
| 69117 | T | ||||||||||||
| 106369 | |||||||||||||
| 13791 | Not Specified | ||||||||||||
| 11747 | |||||||||||||
| 13053 | Not Specified | ||||||||||||
| Only Incorrect Protein IDs | |||||||||||||
| 11596 | T | 15 | 28 | 15 | 25 | 1 | 4 | ||||||
| 11128 | T | 14 | 18 | 14 | 17 | ||||||||
T: trypsin
Ch: chymotrypsin
Lys-C: endoproteinase Lys-C
Glu-C: endoproteinase Glu-C
Sub: subtilisin
ProK: proteinase K
Elas: elastase
Asp-N: endoproteinase Asp-N
None: Protein not digested
Protein Identification
Eight of 42 responding labs correctly identified all three CAH2 isoforms with supporting evidence to discriminate between the isoforms plus N-terminal acetylation of the proteins. One lab correctly identified three isoforms but found N-terminal acetylation on only two of the isoforms. Fifteen of 42 labs correctly identified two of the CAH isoforms with supporting evidence. Eleven of these labs also identified N-terminal acetylation. Nine of 42 labs correctly identified two CAH isoforms, but also made one or more incorrect protein or post-translational modification identifications. Interestingly, four of these labs (#2115, #10567, #11735, and #48583) identified a form of human CAH (CAHIi) that contains a single amino acid substitution for an x-ray crystallography study. Only one lab (#48583) provided an MS/MS spectrum as evidence for this identification. Though unlikely, we cannot rule out the presence of this human isoform in the commercial form of CAH used in this study, so it is possible that these four labs correctly identified three isoforms despite missing one of the bovine isoforms that we know were present (Figures 3a and 3b). Another lab (#11010) identified horse CAH on the basis of the peptide sequence GERQSPVDIDTK, preceded by N in bovine but K in horse. This entry was accompanied by a convincing MS/MS spectrum as supporting evidence that presumably resulted from a “non-tryptic” cleavage after overnight digestion of the sample with trypsin (personal communication from the study participant) that coincidently would have resulted from a canonical tryptic cleavage of the horse isoforms. These examples illustrate the difficulties in accurate discrimination of protein isoforms even when sound methodology and techniques are applied. Seven of 42 labs did not identify any proteins. Two of 42 labs did not make any correct protein identifications, but made one or more incorrect identifications. A total of 33 of 42 labs identified human and bovine isoforms of CAH2. Twenty-seven of 42 labs reported identification of the N-terminus of at least one isoform, while only two of 42 labs reported observing the C-terminus. One laboratory attempted to identify the intact proteins by Edman sequencing, but did not obtain any results, presumably due to N-terminal acetylation of the proteins.
Perception of Difficulty
It was interesting to note that most of the participants felt that they spent the right amount of time working on the sample, though the actual amount of time varied over a wide range. Only about 20% spent more then they intended to, and about 12% spent less then they expected. In actual hours, this compares to 10% spending less than 4 hours, 36% 5 to 8 hours, 22% 8 to 16 hours, and 24% more then 17 hours. Comparing this to the difficulty of the sample, 55% considered it somewhat difficult while 28% considered it fairly straightforward. 2% thought it was easy, while 8% thought it was very difficult. The labs also indicated that 28% did this kind of analysis frequently, while 50% did it only occasionally. Regarding experience, only 7% considered themselves highly experienced, while 50% fell into the experienced category, with 31% somewhat experienced and 2% with no experience. It would appear that the evaluation of difficulty reflects appropriately the amount of time expected to be spent on the sample as well as the experience levels.
Other Survey Responses
In response to the survey request, “Please describe the nature of your difficulties,” three respondents reported that they were unable to detect peptides after trypsin digestion and mass spectrometry. One of these concluded that there was no sample in the tube (“Since we handled the sample over the whole procedure in the original tube which was send to us I have only one explanation: There was no protein inside”), one observed only tryptic peptides, and one reported problems with the mass spectrometer used. A fourth respondent, who attempted to sequence peptides by Edman degradation after tryptic digestion of the proteins followed by HPLC of the resulting peptides, observed peaks in the HPLC chromatogram but was unable to obtain sequences for any of the peptides. Finally, we received 30 responses to the following request: “The Proteomics Research Group values your feedback. If you have any final comments on the study or this survey please enter them here.” We have posted these responses on the ABRF PRG Web site at the following URL: http://www.abrf.org/ResearchGroups/Proteomics/Studies/ABRF2004surveysummary_2695.pdf
CONCLUSIONS
Almost all of the labs employed tryptic digestion of proteins followed by tandem mass spectrometry to identify the proteins.
Separating the proteins before analysis did not influence the success rate of the analyses of this simple protein mixture.
For this study, while two of the eight participants that identified the samples correctly used other enzymes in addition to trypsin, the use of enzymes other than trypsin was not useful for most analyses in this case.
In almost all cases, relying on protein identification software (database search engines) alone was not sufficient to make accurate discriminations between very closely related isoforms (i.e., the two bovine isoforms that differed by a single amino acid). In this particular case, database search engines alone were able to identify the human and bovine forms of carbonic anhydrase. Examination of notation in the database entry was also necessary. Specifically, inspection of the features section of the database record for bovine CAH2 reveals the R→Q single amino acid substitution. This is the case for both the SwissProt knowledge base and the NCBI non-redundant database. Careful inspection of raw MS data was usually required to verify the presence and sequence of the single peptide unique to each bovine CAH2 isoform.
Though the sample number was small, in the case of gel separation the type of gel stain seemed to have an effect on the average sequence coverage that was obtained by mass spectrometry, with Coomassie blue leading to best coverage and silver staining to least coverage.
Overall, responding labs were quite successful in this difficult task.
ACKNOWLEDGMENTS
The Proteomics Research Group thanks Isabel Birg of the Max Planck Institute of Psychiatry for 2D-PAGE analysis of the carbonic anhydrase mixture, Dr. Vivekananda Shetty of the NYU Protein Analysis Facility for LC-MS/MS analysis of the sample mixture, and Lora Goodrich of Columbia University for participant correspondence and ensuring anonymity of the participants. We offer special thanks to all of the scientists who took part in this study and submitted results. We acknowledge support from NIH Shared Instrumentation Grant S10 RR017990 to T.A.N.
REFERENCES
- 1.Arnott DP, Gawinowicz M, Grant RA, Lane WS, Packman LC, Speicher K, et al. Proteomics in Mixtures: Study Results of ABRF-PRG02. J Biomol Tech. 2002;13:179–186. [PMC free article] [PubMed] [Google Scholar]
- 2.Arnott D, Gawinowicz MA, Grant RA, Neubert TA, Packman LC, Speicher KD. ABRF-PRG03: Phosphorylation site determination. J Biomol Tech. 2003;14:205–15. [PMC free article] [PubMed] [Google Scholar]