Abstract
We describe a reliable protocol for constructing single-site saturation mutagenesis libraries consisting of all 20 naturally occurring amino acids at a specific site within a protein. Such libraries are useful for structure-function studies and directed evolution. This protocol extends the utility of Stratagene’s QuikChange Site-Directed Mutagenesis Kit, which is primarily recommended for single amino acid substitutions. Two complementary primers are synthesized, containing a degenerate mixture of the four bases at the three positions of the selected codon. These primers are added to starting plasmid template and thermal cycled to produce mutant DNA molecules, which are subsequently transformed into competent bacteria. The protocol does not require purification of mutagenic oligonucleotides or PCR products. This reduces both the cost and turnaround time in high-throughput directed evolution applications. We have utilized this protocol to generate over 200 site-saturation libraries in a DNA polymerase, with a success rate of greater than 95%.
Keywords: site-directed saturation mutagenesis, directed evolution
Directed evolution is a proven and powerful tool for modifying the catalytic abilities of enzymes.1 Improved enzymes are identified in mutant libraries, in some instances screening for incremental improvements through multiple generations of mutation and selection. Practical constraints on library size and screening capacity emphasize the importance of library quality. Mutant libraries are typically constructed using error-prone PCR, DNA shuffling, or regional site-saturation mutagenesis. In this communication, we report simple modifications to the Stratagene QuikChange protocol facilitating the construction of single-site saturation libraries for directed evolution. Such libraries are collections of recombinant clones that contain all 20 naturally occurring amino acids at a specific site in a protein of interest.
In order to improve utilization of novel phosphate-labeled nucleotides by DNA polymerases, we subjected Therminator DNA polymerase (New England Biolabs, Beverly, MA) to five generations of mutation and selection using single-site saturation libraries. A total of 80 amino acid positions were chosen for mutagenesis, based on their proximity to the supposed location of the nucleotide modification in a structural model of the enzyme. Each screened library comprised 93 mutants randomized at one codon, giving a good probability to represent all 20 amino acids. At each of five generations, we selected the most improved mutant polymerase from a collection of up to 40 different libraries (amino acid positions). The obtained new mutation was fixed, and unselected amino acid positions were again targeted for mutagenesis in the next generation. Overall, we constructed over 200 libraries and screened over 65,000 individual mutants. Enzyme activity improved some 40-fold, until the incorporation rate of the modified dNTPs reached parity with natural dNTPs at about 20 nucleotides per sec.
Many methods for mutagenesis have been described.2–11 We constructed site saturation libraries using Stratagene’s single-site QuikChange Site-Directed Mutagenesis Kit. While commonly used to change single codons and to insert or delete DNA segments (http://www.stratagene.com/manuals/200523.pdf), this kit is not recommended for degenerate (one-codon) mutagenesis. The latter application is suggested only with a more elaborate kit, the QuikChange Multi Site-Directed Mutagenesis Kit.5 In addition, both kits recommend HPLC- or gel-purified mutagenic primers. This additional purification is significant both in time and cost when used in a high-throughput setting. We show here that the simpler single-site Quik-Change Kit is sufficient for constructing degenerate one-codon libraries, and that mutagenic oligonucleotides from Operon (Huntsville, AL) can be used desalted with no further purification.
METHODS, RESULTS, AND DISCUSSION
The codon to be randomized is chosen and two complementary oligonucleotides are synthesized corresponding to the two DNA strands. Equimolar degenerate mixes of all four bases (A, T, G, and C) are incorporated at the three positions of the selected codon. Each arm flanking the degenerate site should be approximately 15–20 nucleotides in length and contain a minimum of six Gs and/or Cs. The sequence of the mutagenic primers is dictated by the DNA sequence of the region to be mutated—only the primer’s beginning and ending points can be altered. Total primer length is usually 30–40 bases. Ideally, the G+C content should be 45–55%, and TM in the range of 70–95ºC. There should also be no self-complementary palindromic sequences on either the 3′ or 5′ end of the primers to avoid primer-dimer formation. If possible, avoid highly stable hairpin loops. Desalted primers are dissolved in 0.1X TE (1 mM Tris, 0.1 mM EDTA, pH 8.0) and diluted in deionized H2O to 2 μM concentration.
For starting template, we utilized a recombinant plasmid (approximately 6.5 kb) isolated using a QIAGEN QIAprep Spin Miniprep Kit (Valencia, CA). This methodology requires methylated parental DNA, which is the case for most commonly utilized E. coli strains. Mutagenesis reactions were performed in a 25-μL volume containing 1X Pfu reaction buffer (10 mM KCl, 10 mM (NH4)2SO4, 20 mM Tris-HCl (pH 8.8), 2 mM MgSO4, 0.1% Triton X-100, and 0.1 mg/mL BSA), 20 ng template plasmid DNA, 6 pmol of each primer, 200 μM of each dNTP, and 1 unit of PfuTurbo DNA polymerase (cloned Pfu and PfuUltra Hotstart [Stratagene, La Jolla, CA] have also been used successfully). Often, multiple reactions were performed simultaneously, in which case a master mix was assembled on ice, then aliquoted into individual tubes, each containing the appropriate primers. Reactions were thermal cycled: 95ºC for 2 min, followed by 16 cycles of 95ºC for 30 sec, 55ºC for 1 min, and 68ºC for 10 min, then a final incubation of 68ºC for 10 min. Larger plasmids may require longer extension times. Reactions were cooled on ice and digested with 5 units of Dpn I for 1 h at 37ºC to cleave methylated and hemimethylated parental DNA, but not the newly synthesized mutant DNA molecules. Completed reactions were either cooled on ice or stored at –20ºC. Another publication has reported using QuikChange methodology to generate site-saturation libraries.11 However, we have not found it necessary to use partial overlapping primers, purified mutagenesis primers, or purified PCR reactions as they did.
Reactions were used to transform 50 μL of chemically competent TOP10 E. coli (Invitrogen, Carlsbad, CA). Cells were thawed on ice, 5 μL of reaction added, mixed gently, then incubated on ice for 30 min. Cells were heat-shocked at 42ºC for 30 sec, then placed on ice again for 1 2 min. Next, 250 μL of SOC media (Sigma Chemical, St. Louis, MO) was added, and transformed cells incubated at 37°C with shaking for 1 h. Finally, 100 150 μL was plated onto LB agar plates containing ampicillin (100μg/mL) and incubated overnight at 37ºC. Generally, 100–500 colonies appeared on the plate the next day and were ready for further analysis. Our success rate has been over 95%. Failures are usually an insufficient number of clones appearing on the plate. This situation can frequently be rectified by plating more of the transformation mixture or repeating the transformation.
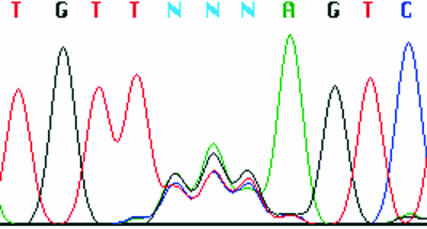
Plasmid DNA has been isolated from these saturation libraries using a QIAGEN Miniprep Kit (Valencia, CA) and analyzed by DNA sequencing. Figure 1 shows one selected randomized site. Approximately equal quantities of the four bases can be seen at each position of the codon. This was not the case when mutagenesis reactions were electroporated into cells. Here, a larger proportion of wild-type bases was present. The reason for this is unclear, but may be due to the presence of intact undigested parental plasmid, which may be more efficient at electroporating cells than newly synthesized, unligated mutant DNA molecules.
FIGURE 1.
DNA sequencing of a site-directed mutagenesis library. DNA was isolated from the library using a QIAGEN QIAprep Spin Miniprep Kit (Valencia, CA), and sequenced using an infrared dye–labeled primer on a LI-COR Model 4200 Automated Sequencer (Lincoln, NE). The randomized three positions of the codon are clearly visible.
ACKNOWLEDGMENTS
The authors wish to thank Jon Anderson, Teresa Urlacher, and Lyle Middendorf for critical reading of the manuscript. This work was supported by grants from the NIH/NHGRI (R43 HG002292, R44 HG002292, and PO1 HG003015). The authors declare that they have no competing interests.
REFERENCES
- 1.Arnold FH, Georgiou G, editors. Methods in Molecular Biology. Vol. 231. Totowa, NJ: Humana Press; 2003. Directed Evolution Library Creation, Methods and Protocols. [Google Scholar]
- 2.Adereth Y, Champion KJ, Hsu T, Dammai V. Site-directed mutagenesis using Pfu DNA polymerase and T4 DNA ligase. BioTechniques. 2005;38:864–868. doi: 10.2144/05386BM03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angelaccio S, di Patti MCB. Site-directed mutagenesis by the megaprimer PCR method: Variations on a theme for simultaneous introduction of multiple mutations. Anal Biochem. 2002;306:346–349. doi: 10.1006/abio.2002.5689. [DOI] [PubMed] [Google Scholar]
- 4.Gong Z, Zhang H, Gabos S, Li X. Rapid and efficient polymerase chain reaction–based strategies for one-site and two-site mutagenesis. Anal Biochem. 2004;331:404–406. doi: 10.1016/j.ab.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 5.Hogrefe HH, Cline J, Youngblood GL, Allen RM. Creating randomized amino acid libraries with the QuikChange Multi Site-Directed Mutagenesis kit. BioTechniques. 2002;33:1158–1165. doi: 10.2144/02335pf01. [DOI] [PubMed] [Google Scholar]
- 6.Miyazaki K, Takenouchi M. Creating random mutagenesis libraries using megaprimer PCR of whole plasmid. BioTechniques. 2002;33:1033–1038. doi: 10.2144/02335st03. [DOI] [PubMed] [Google Scholar]
- 7.Nagy ZB, Felföldi F, Tamás L, Puskás LG. A one-tube, two-step polymerase chain reaction–based site-directed mutagenesis method with simple identification of the mutated product. Anal Biochem. 2004;324:301–303. doi: 10.1016/j.ab.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 8.Sawano A, Miyawaki A. Directed evolution of green fluorescent protein by a new versatile PCR strategy for site-directed and semi-random mutagenesis. Nucleic Acids Res. 2000;28:e78. doi: 10.1093/nar/28.16.e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei D, Li M, Zhang X, Xing L. An improvement of the site-directed mutagenesis method by combination of megaprimer, one-side PCR and DpnI treatment. Anal Biochem. 2004;331:401–406. doi: 10.1016/j.ab.2004.04.019. [DOI] [PubMed] [Google Scholar]
- 10.Wong TS, Tee KL, Hauer B, Schwaneberg U. Sequence saturation mutagenesis (SeSaM): A novel method for directed evolution. Nucleic Acids Res. 2004;32:e26. doi: 10.1093/nar/gnh028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng L, Baumann U, Reymond J. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004;32:e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]