Abstract
Microarrays have revolutionized many areas of biology due to our technical ability to quantify tens of thousands of transcripts within a single experiment. However, there are still many areas that cannot benefit from this technology due to the amount of biological material needed for microarray analysis. In response to this demand, chemistries have been developed that boast the capability of generating targets from nanogram amounts of total RnA, reflecting minimal amounts of biological material, on the order of several hundred or thousand cells. Herein, we describe the evaluation of four chemistries for RnA amplification in terms of reproducibility, sensitivity, accuracy, and comparability to results from a single round of T7 amplification. No evidence for false-positive measurements of differential expression was observed. In contrast, clear differences between chemistries in sensitivity and accuracy were detected. PCR validation showed an interaction of probe sequence on the array and target labeling chemistry, resulting in a chemistry-dependent probe set sensitivity varying over an order of magnitude.
Keywords: microarray, gene expression, RNA amplification, small sample, GeneChip, quantitative PCR
Gene expression profiling is a powerful method to gain an in-depth understanding of a cell’s transcriptional response to external stimuli. Micro-arrays have evolved to become the technology of choice for expression profiling, allowing for broad surveys of tens of thousands of genes in a single experiment. Over the past five years, methods for creating highly reproducible and standardized microarrays have aligned with improvements in experimental design and execution to produce a technical platform that promises to be useful beyond the laboratory and into the clinic.1–3
While the growth of this technology has been impressive, technical limitations have slowed even more rapid progress. For instance, until recently, microarrays have required relatively large quantities (5–10 μg) of total cellular RNA as a target labeling substrate.4,5 This requirement has hampered the incorporation of microarrays in the clinic, where biopsies are precious and procedures must be minimally invasive. Further, parallel advancements in technologies such as laser capture microdissection (LCM)6 have revealed the utility of further dissecting these small samples to even smaller histological substructures for expression analysis.3,7 This has resulted in the availability of only nanogram quantities of total RNA for each sample, creating a demand for technologies that will provide sufficient target for expression profiling without introducing excessive bias in transcript representation.
In response to this demand, a number of target amplification technologies have been developed and optimized,8–11 and several of these have been made commercially available.
To evaluate the relative merits of several of these technologies, four commercial target amplification protocols were investigated and compared to the standard target labeling procedure using the Affymetrix GeneChip platform. The commercial small sample target synthesis protocols evaluated include:
Ovation (Nugen Technologies, Inc., San Carlos, CA)
Message Amp (Ambion, Inc., Austin, TX)
Small Sample Target Labeling Assay Version II (Affymetrix, Inc., Santa Clara, CA)
BioArray Small Sample Amplification Protocol (Enzo Life Sciences, Inc., Farmingdale, NY)
A total of six samples were labeled for each technology (triplicates of two different human RNA samples). Five micrograms of total RNA was labeled using the standard protocol and 10 ng of total RNA was labeled using the small-sample protocols described above. In addition to data quality (bias, noise, reproducibility, etc.) and comparison of the small sample protocols to the standard protocol, an evaluation of the technical merits of each procedure (ease of use, scalability, automatability, etc.) was made. The results of this systematic evaluation have provided useful insight for investigators attempting to determine which procedure will work best in their laboratory. This study will also present the commercial providers a real-world evaluation of their technical platforms, allowing for continued improvements in the field of target amplification.
The study described herein addresses a number of questions: How reproducible are the measurements of gene expression for each of the amplification chemistries (within each technology)? Are there differences in sensitivity for differential expression between small sample amplification approaches? How comparable are the results generated by amplification compared to the results generated by a standard single-round amplification protocol? What is the degree of accuracy for measurements after amplification—for example, how well does measured differential expression reflect true differences in amounts of transcripts between samples? Lastly, how similar or different are biological conclusions drawn from results of each of the chemistries?
These questions and others have been raised, tested and addressed in this study. Although gene expression is measured on a relative scale, it is important to evaluate the consistency and performance of amplification chemistries as a function of sample quality and biological relevance.
MATERIALS AND METHODS
RNA Extraction, Quantification, and Quality Control
Total RNA was extracted by Trizol (Invitrogen, Carlsbad, CA) from a breast adenocarcinoma cell line (MDA-MB-231, ATCC) or obtained from Stratagene (Human Reference RNA). After a cleaning step on RNeasy column (Qiagen, Valencia, CA), each sample was aliquoted at 0.5 μg/μL for the standard protocol or 10 ng/μL for the small sample preparation protocols. RNA concentrations were calculated using a Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies, Rockland, DE). Riobosomal subunit intactness (a correlative measure that indicates intact mRNA) was ensured before labeling by analyzing 20–50 ng of each sample using the RNA 6000 NanoAssay for the Bioanalyzer 2100 (Agilent). Both samples had a 28S/18S ribosomal peak ratio between 1.8 and 2.0. Each sample was amplified and biotin labeled in triplicate following five different protocols (the standard protocol and four different small-sample preparation protocols).
Logistics
In order to reduce environmental or nonbiological variations, the samples were handled and processed by the same technician from beginning to end. The five labeling protocols were performed in the same laboratory (Genomics Core Lab-MSKCC), and triplicates for both samples were run in parallel for a given protocol. All samples were processed over a two-week period.
Standard Protocol
The standard protocol described by Affymetrix at https://www.affymetrix.com/support/downloads/manuals/expression_s2_manual.pdf was used for one round T7 amplification. Five micrograms of total RNA was used for cDNA synthesis using an oligo-dT-T7 primer and the SuperScript Double-Stranded cDNA Synthesis Kit (Invitrogen). Synthesis, linear amplification, and labeling of cRNA were accomplished by in-vitro transcription using the Bioarray High Yield RNA Transcript Labeling kit (Enzo Life Sciences).
Small Sample Preparation Protocols
We used the following kits to amplify and label 10 ng of total RNA for both samples: Affymetrix two-cycle target labeling protocol (Cat. No. 900494), Enzo Life Sciences BioArray RNA Amplification and labeling system (Cat. No. 42410S-2B), Ambion MessageAmp II aRNA kit (Cat. No. 1751), and NuGEN Ovation biotin system (Cat. No. 2300-12). Each protocol was strictly followed as described by the manufacturer. Detailed procedures can be downloaded from the ABRF Microarray Research Group Web site at http://www.abrf.org/index.cfm/group.show/Microarray.30.htm#R_4.
Microarray Hybridization Image Processing
Fifteen micrograms of labeled and fragmented cRNA (Standard protocol, Affymetrix, Enzo and Ambion small-sample protocols) or 2.2 μg of cDNA (NuGEN protocol) were then hybridized to the Human Genome U133A2.0 GeneChip (Affymetrix), which contains 22,215 oligonucleotide-based probe sets, at 45°C for 16 h.
Post-hybridization staining and washing were done according to manufacturer’s recommendations (Affymetrix). Finally, chips were scanned in the GS3000 scanner (Affymetrix). Images were converted to CEL files using GCOS1.1 (Affymetrix).
Real-Time PCR
Two micrograms of total RNA were reverse-transcribed using the iScript cDNA Synthesis kit (Biorad, Hercules, CA) at 42°C for 30 min. Forty nanograms of resultant cDNA was used in a Q-PCR reaction using an iCycler (Biorad) and pre-designed TaqMan Gene expression assays (Applied Biosystems). Primers were chosen based on their ability to span the most 3′ exon-exon junction. Amplification was carried out for 40 cycles (95°C for 15 sec, 60°C for 1 min). To calculate the efficiency of the PCR reaction and to assess the sensitivity of each assay, we also performed a seven-point standard curve (10, 3.3, 1.1, 0.37, 0.123, 0.041, and 0.015 ng) using cDNA. Triplicate CT values were averaged, and amounts of target were interpolated from the standard curves and normalized to hypoxanthine guanine phosphoribosyl transferase gene expression.
Data Analysis
Analytical procedures focused on defining the influence of RNA labeling protocols on microarray results using Affymetrix U133A2.0 platforms. Two RNA samples (MDA and RR) were labeled with five different protocols (Affymetrix [Af], Ambion [Am], Enzo [En], Nugen [Ng], and standard [Std]) in triplicate generating 15 CEL files for each RNA sample. Expression summaries were generated for each probe set from each CEL file with Robust Multichip Analysis (RMA). RMA is implemented in the following manner: (a) probe-specific correction of the PM probes using a model based on observed intensity being the sum of signal and noise; (b) normalization of corrected PM probes using quantile normalization12; (c) calculation of expression measure using median polish. The correlation of the probesets to each other for each hybridization was done in Excel. The correlation coefficient for every pairwise comparison of each labeling protocol for a given RNA sample was done, tabulated, and is contained in the associated Excel spreadsheet. Data have been posted in the NCBI GEO database (http://www.ncbi.nlm.nih.gov/geo/) and can be retrieved using the following access number: GSE2723.
RESULTS
Reproducibility
For each of the chemistries, three aliquots of each of the RNA samples have been processed from amplification to scanning of microarrays, resulting in six results files. Three correlation coefficients (r2) have been calculated for pairwise comparisons between expression estimates of replicates of each chemistry and the mean of r2 values has been calculated. For all four amplification chemistries as well as for the standard protocol, average r2 values are above 0.99, indicating high reproducibility of measurements.
The global high intra-chemistry correlation could potentially lead to confounding interpretations, because varying chemistries leads to artificial difference in sensitivity to noise at decreasing signal levels. To examine this possibility, we evaluated intra-chemistry correlation by signal intensity quartile. Figure 5 displays the dependency of intra-chemistry correlation on signal intensity quartile. As the signal intensity decreases, there is a general decrease in intra-chemistry correlation even for the standard chemistry, as one would expect given the Monte Carlo effect. All the commercial small-sample amplification protocols fall slightly below the standard at the bottom quartile except NuGEN’s Ovation, which falls about 15% below the standard. The difference in noise (as defined by specific signal intensity as a function of complementary hybridization between the amplification technologies does not seem to reflect output metrics as described below (Figure 5).
FIGURE 5.
Intra-chemistry correlation by signal intensity. Probe sets were divided into quartiles based on average signal intensity for each chemistry. Six correlation measurements for each chemistry at each quartile were based on three RR samples compared to themselves and three MDA samples compared to themselves. These six correlations were averaged and the coefficient of variation was computed. The coefficient of variation for any average did not exceed 5%.
Sensitivity
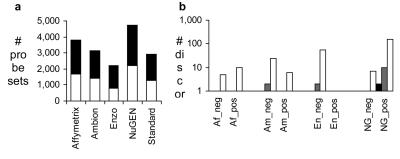
For each of the chemistries, differential expression values were calculated from mean expression estimates per set of replicates. The number of probe sets measuring more than a twofold ratio of tumor vs. reference RNA differential expression is shown in Figure 1a. Results from samples processed using the NuGEN chemistry showed the highest number of differentially expressed probe sets, followed by Affymetrix, Ambion, and Enzo. The standard procedure measured a number of differentially expressed genes, between Ambion and Enzo results.
FIGURE 1.
Sensitivity of labeling chemistries to detect differential expression and comparability of results to the standard protocol. (a) Number of probe sets detecting >2-fold differential expression; white bars show probe sets up-regulated in MDA, black bars show down-regulated. (b) Sensitivity discordances in measurements of differential expression between amplification chemistries and standard protocol; a discordance between an amplification chemistry and the standard protocol was counted when one chemistry measured strong differential expression (black bars >64-fold, gray bars >16-fold, white bars >4-fold) while the other chemistry measured < 1.5-fold differential expression. A discordance is counted as negative (e.g., NG_pos for NuGEN) when the amplification chemistry measured strong differential expression while the standard protocol gave < 1.5-fold differential expression.
Comparability of Amplification Results to Results of Standard Procedure
For a global picture of similarity of results between different chemistries, absolute expression estimates (as defined by the number of differentially expressed genes across chemistries) were compared. For both samples, expression estimates from each of the amplification chemistries was compared to expression estimates of each of the replicates of the corresponding sample processed according to the standard procedure. Results of samples processed by Affymetrix chemistry showed the highest correlation to standard procedure, followed by Enzo, Ambion, and NuGEN (Table 1).
TABLE 1.
Comparison of Results from Amplification Chemistries to Standard Procedure
| Correlation to Standard Protocol (r2)a | Directional Conflicts >2-fold | Directional Conflicts >1.5-fold | |
|---|---|---|---|
| Affymetrix | 0.96 | 0 | 0 |
| Ambion | 0.89 | 0 | 1 |
| Enzo | 0.94 | 0 | 0 |
| NuGEN | 0.84 | 0 | 11 |
For both samples RR and MDA, expression estimates from each of the amplification chemistries was compared to expression estimates of each of the replicates of the corresponding sample processed according to the standard procedure. Mean r2 values from nine pair-wise comparisons are shown for each chemistry. Number of probe sets showing directional conflicts between differential expression measured by the standard protocol and each of the amplification chemistries is shown. A directional conflict is counted as >1.5-fold if the standard protocol measures differential expression >1.5-fold (log2 difference absolute >0.58) while the indicated amplification chemistry measures opposite direction of differential expression >1.5-fold. The same principle holds true for directional conflicts >2-fold (log2 difference absolute >1).
As a next step, differential expression between samples RR and MDA was calculated from measurements using amplification chemistries as well as using the standard procedure. Directional conflicts were calculated between standard procedure and each of the amplification chemistries, where a directional conflict was defined as follows: Standard procedure measures higher (lower) expression in RR than in MDA while an amplification chemistry measures the opposite relation (Table 1). No probe sets shows greater than twofold directional conflicts between chemistries.
To characterize quantitative differences in measurements of differential expression using the different chemistries, sensitivity discordance was determined. Discordance between sensitivity of amplification chemistries and the standard procedure was defined as follows: An amplification chemistry measures strong differential expression (over 4-, 8-, or 16-fold) while the standard procedure measures minor differences (under 1.5-fold, i.e., 50% up or 33% down-regulation) and vice versa. Enzo, the chemistry showing the lowest sensitivity for differential expression, generates the highest number of negative discordances. A negative discordance is defined as minor differences measured by the amplification chemistry while the standard procedure measures strong differential expression. NuGEN, the chemistry showing the highest sensitivity, highlights the highest number of positive discordances. Up to 64-fold differential expression is measured by NuGEN chemistry when the standard procedure measures differences under 0.5-fold changes from the same probe set.
Accuracy of Measurements of Differential Expression
To find out how far measurements of differential expression represent real differences between samples, micro-array measurements were compared to measurements by real-time PCR. For validation, transcripts were identified where T7 RNA polymerase–based chemistries (Standard procedure, Affymetrix, Ambion, and Enzo) generated results very different from results of isothermal amplification (NuGEN), since NuGEN results showed the highest rate of discordance compared with the all other chemistries. Five transcripts were validated by real-time PCR; NuGEN identified strong differential expression (>2-fold) while the standard procedure measured minor differences between samples (< 1.5-fold); in four transcripts, NuGEN measured minor differences while the standard procedure measured strong differential expression according to the criteria above. In all cases, real-time PCR measured strong differential expression (Figure 2a).
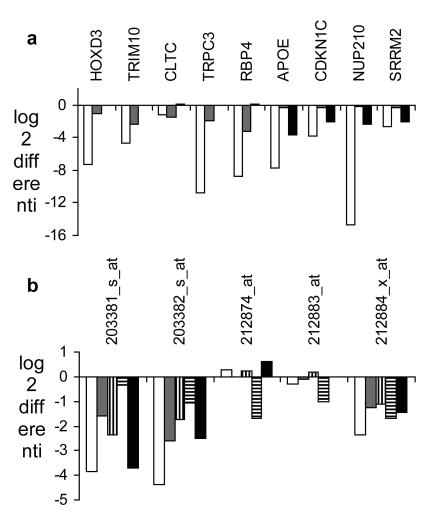
FIGURE 2.
Confirmability of array results by qPCR and internal consistency of array results. (a) Differential gene expression measured by qPCR (white bars), by NuGEN (gray bars) and by the standard protocol (black bars) for nine transcripts where at least one probe set indicated sensitivity discordance between the two chemistries. (b) Measurement of differential expression of ApoE by five probe sets (white bars show results from Affymetrix chemistry, gray bars from Ambion, diagonally striped bars from Enzo, horizontally striped bars from NuGEN, and black bars from standard protocol chemistry).
As another line of evidence for accuracy of measurements, internal consistency was used between multiple probe sets measuring the same transcripts. In many cases, more than one probe set per transcript is present on the U133A microarray. Since PCR measurements confirmed strong differential expression in all cases tested, we hypothesize that insensitivity (false-negative measurements) is a bigger problem than false-positive results for differential expression. Based on this hypothesis, if at least two chemistries measured strong differential expression on at least one probe set, we assumed that this transcript is truly differentially expressed. For eleven transcripts, at least three probe sets were available on the U133A2.0 microarray, and in six cases at least two chemistries detected strong differential expression of the same transcript using at least two probe sets. An example is shown for apolipoprotein E (ApoE, Figure 2b). Using two probe sets, all chemistries detected strong differential expression; using one probe set, four out of five chemistries detected strong differential expression; and on two probe sets, only isothermal amplification pointed in the same direction. Real-time PCR confirmed these findings of strong differential expression of ApoE. Another example for chemistry-dependent sensitivity of probe sets is Nucleoporin 210 (NUP210), where two probe sets showed quantitative discordance within results of isothermal amplification as well as within the standard procedure, but the discordance points into the opposite direction for the chemistries. Raw fluorescence intensities of probes (Figure 3) within the two probe sets point to true differences in probe sensitivity instead of potential artifacts generated by data analysis. Real-time PCR confirmed the finding of strong differential expression of NUP210 (Figure 2a). From the results of real-time PCR as well as the measurements of the same transcript by multiple probe sets, we do not see evidence of false-positive results of differential expression.
FIGURE 3.
Raw fluorescence intensities show chemistry-dependent probe sensitivities. Fluorescence intensities of all 11 probes of probe sets 213945_s_at (upper panel) and 220035_at, both measuring NUP210. Triangles show signal intensity of RR and squares of MDA; (a) and (c) signals from NuGEN chemistry, (b) and (d) from standard protocol.
Biological Interpretation of Results
To learn about pathway-based results generated by the different chemistries, functional analysis was performed on results from each of the chemistries. Pathways were identified (using PathwayAssist; Iobion Informatics), where strongly differentially expressed gene products were correlated using gene function information in the public domain. A comparison of results from the chemistry showing highest sensitivity (NuGEN, measuring the highest number of differentially expressed transcripts) to the chemistry showing lowest sensitivity (Enzo) was performed (Figure 4). The same pathways were identified, but the number of interacting gene products was higher for results of isothermal amplification.
FIGURE 4.
Pathway analysis was performed on the best- and worst-performing protocols to determine whether the biological implications of differentially expressed genes was compromised. The most differentially expressed genes from the Enzo two-round and NuGEN Ovation protocols were used in this comparison. PathwayAssist was used to generate direct (and indirect) interactions of differentially expressed genes between the two processing protocols. The gene products circled in blue represent hits from the Enzo analysis and demonstrate that the core biological mediators in this analysis are the same when comparing differentially expressed genes between NuGEN and Enzo. It is also clear that the NuGEN approach (in this comparison) yielded a significant increase in the number of genes identified in the pathways that correlate to the core sets of genes identified by both approaches. The main point is that even in the absence of a true biological model (as in this comparison with a reference gene) the core set of biologically interacting genes in this pathway are preserved between both small-sample amplification methods; however, a more sensitive amplification approach can lead to the detection of a larger set of differentially expressed genes within a pathway.
Practical Metrics and Performance Criteria
There are other important factors when evaluating small-sample amplification protocols that laboratories should consider. These include metrics such as time, technical effort, portability, etc. These metrics were measured during the training and protocol execution process and described in Table 2. Interestingly, there is a significant difference among some protocols, and not surprisingly the time to completion is directly correlated to the number of steps needed in each protocol. Although the protocols performed in a comparable manner, data such as processing time can potentially factor into a lab’s decision to adopt an approach based on workflow of the group. With proper training (in some instances by the manufacturer) the protocol performance is high, and all cases met the manufacturers’ published performance metrics.
TABLE 2.
Performance and Practical Metrics of Small-Sample Amplification Protocols
| A | ||||||
|---|---|---|---|---|---|---|
| Protocol | RNA Amt. (nanograms) | Amp yield cRNA/cDNA* | Background | GAPDH 3′/5′ – 1.1Kb | β–Actin 3′/5′—1.8 Kb | % Present |
| 2x Enzo | 20 | 80 | 80 | 1.2 | 17.8 | 49% |
| 2x Affy | 20 | 149 | 61 | 1.7 | 17.7 | 60% |
| 2x Ambion | 20 | 102 | 49 | 8.5 | 23.7 | 54% |
| NuGEN | 20 | 5* | 38 | 1.9 | 25.8 | 65% |
| B | ||||||
| Protocol | Protocol Time (to arrays) | Labor Intensity | Technical Support | List Price (all inclusive) | ||
| 2x Enzo | 2.5 d | 3 | 3 | $231 | ||
| 2x Affy | 2.5 d | 2 | 2 | $153 | ||
| 2x Ambion | 2.5–3.0 d | 2 | 1 | $143 | ||
| NuGEN | 1–1.5 d | 1 | 1 | $194 | ||
(A) Performance metrics for small-sample amplification technologies. These metrics are standard metrics employed by investigators to assess and compare array performance prior to biological evaluation.
(B) Practical metrics of small-sample amplification performance was measured. A score of 1 is considered the best performance, while a score of 5 is considered worst performance.
The two best performing small-sample protocols as a function of this evaluation (i.e., reproducible results, sensitivity, validation accuracy, ease of use, data portability) are the Affymetrix two-round amplification protocol and the NuGEN Ovation small-sample amplification technology. It is also important to note that although the amplification performance and array metrics were similar, the actual genes within each technology differed in some instances within and between amplification approaches (Figure 6). For the most part, the core set of differentially expressed genes was conserved (Figure 4), but additional targets within the same biological framework are uncovered by the approaches that ended up being assessed as more reproducible, specific, and sensitive.
FIGURE 6.
Hierarchical clustering of all hybridization groups. All genes were clustered using a Pearson correlation following a filter based on signal intensity (100 cutoff). All samples clustered correctly within amplification technology, and the genes highlighted by the yellow box constitute the roughly 1500 additional differentially expressed genes in NuGEN amplified samples. Closer evaluation of all hybridization groups shows important differences between technologies, but most importantly demonstrates the overall comparability (on a gene-by-gene level) of all technologies evaluated. Std: 1x Affy; En: 2x Enzo; Af: 2x Affy; Am: 2x Ambion; Ng: NuGEN Ovation; RR: reference RNA; MDA: tumor RNA.
DISCUSSION
Small-sample amplification is a critical component to all microarray technologies with the continued refinement of both basic science and clinical protocols that require high throughput gene expression. With a variety of approaches to choose from it is of paramount importance to understand the differences in reproducibility and efficiency with commercially available technologies prior to biological inquiry. To date most approaches for small sample amplification have been modifications of the more standard T7 in vitro transcription protocol9 adapted to accommodate multiple rounds of amplification in order to generate target from small amounts of RNA. The study described herein asked many questions regarding sample processing that will be useful for the community when planning new experiments where sample is limited. In addition, this body of work addresses the importance of data analysis at the probe level for traditional single-round amplified samples as well as small-sample amplification protocols.
One of the first metrics used for assessing sample fidelity following amplification is transcript size (i.e., message distribution). This is often performed by gel electrophoresis or, more recently, using a variety of lab-on-a-chip technologies (i.e., Agilent Bioanalyzer 2100). This metric is used in combination with yield of cRNA or cDNA to provide a first estimate of sample fitness prior to array hybridization. Although this metric is important when looking at within-sample fidelity, it is of less use when comparing sample reproducibility from a biological perspective, because it does not reveal any information about the relative contribution of transcripts in the population. In addition, the experimentalist needs to look for sample-to-sample consistency and not overall transcript size, because this is a function of the individual laboratory’s performance and not a metric that leads to comparability across laboratories. Lastly, it is important to note that most commercially available microarray technologies contain a rather severe 3′ bias in their oligonucleotide designs and greater than 96% of all Affymetrix probe sets are designed to regions less than 700 bp from the 3′ end of the gene. Therefore, the median length of amplified transcripts ±100 bp from this target will be ensured to cover content represented on the array. The average fragment lengths achieved in this study are within the specifications needed to provide optimal performance on Affymentrix GeneChips and are depicted in Figure 7. The hybridization kinetics and fragment size (i.e., reproducibility) play a large role in any of the amplification technologies, as is discussed in detail below. All of the technologies evaluated exceed the criteria described above with regard to within-technology reproducibility and also meet the length criteria with respect to the Affymetrix technology being utilized in this study. In light of these two pieces of information, the hybridization analysis is more straightforward, since no allowances need to be made for any technology that is not fit to represent all genetic content on the arrays.
FIGURE 7.
3′ bias of Affymetrix Genechip Arrays—Affymetrix probe arrays (human U133 array depicted here) represent a severe 3′ bias which has a direct effect on array peformance as a function of the “size” of labeled product being hyrbridized to the array. Labeled target sequences over 600 bp represent a very small number of Affymetrix transcripts on any given array.
One of the central issues addressed across technologies evaluated is related to hybridization. Items that affect hybridization include (and are not limited to) fragment size, hybridization cocktail buffer, probe nucleic acid and substrate, and target nucleic acid being hybridized. With respect to the first three issues, the different small-sample technologies all utilize the same metrics (i.e., buffer composition, fragmented target size range), normalizing any potential differences that would lead to hybridization differences. With that said, one small-sample protocol has a fundamental difference in the target nucleic acid in the hybridization cocktail. The NuGEN amplification technology is a cDNA amplification approach, which yields a single-stranded cDNA target in contrast to the T7-based protocols, generating cRNA. In theory, cDNA hybridization might be expected to be more specific due to the hybridization kinetics of a DNA-DNA interaction when compared to the higher energy RNA-DNA hybrid, which may lead to less specific interactions due to the inability to effectively dissociate nonspecific interactions.4 Indeed, the data set showed (at the individual probe level) more specific binding via better correlation to multiple probe sets per gene. However, some discordance still remained between cRNA and cDNA hybridization yielding not better or worse results but additional findings between the two fundamentally different technologies. The concordance between cRNA and cDNA hybridizations can be determined by looking at the individual differentially expressed genes; however, the overall concordance between individual groups is most interesting. The number of additional targets identified using the NuGEN technology increased from 3600 to 4800 when compared to the best two-round cRNA approach. Certainly, more detailed validation of a larger number of these targets, by quantitative PCR, would be needed to support more in-depth conclusions about data comparability; however, all of the genes that were expressed in one technology exclusively validated by QPCR. Lastly, the differences that are observed are also slightly skewed in that fold change comparisons may or may not include a gene if it misses a pre-defined cutoff. With that said, the single-primer isothermal linear amplification produced a cDNA target that led to a more robust result than comparable T7-based approaches. This is evident in Figure 4, where the number of partners in a given functional pathway is increased in the NuGEN amplified samples. The discordant differentially expressed genes (11 probes out of 23,000) were not validated and represent 0.04% of the entire population queried. It is important to note that all of the genes differentially expressed by T7 but not recognized by NuGEN also validated, albeit with a smaller number of targets in the discovery process. Studies are still required to fully validate the accuracy of the magnitude of change between the different technologies (and technology providers offering the same approach); however, preliminary analysis points towards cDNA hybridization as being more accurate in this regard.
Ultimately, investigators will want to compare data across different microarray platforms. Although this challenge presents unique hurdles, more immediately the community would be in more effectively comparing data across laboratories (protocol reproducibility) and across experiments (protocol comparability). The former was measured by looking at the microarray output metrics within a technology across the samples being evaluated. For all technologies utilized, the reproducibility for replicates within an experiment measured r2 = 0.99, leading to the conclusion that a properly trained and technically experienced lab can achieve a level of reproducibility for all technologies evaluated that should allow for effective data comparison across laboratories utilizing the same technology. This makes the assumption that sample origin and quality across institutions making the comparison are extremely reproducible in their own right. This is an assumption that is often overlooked and leads to results seen in another MARG study (unpublished observation), where 5000 Affymetrix arrays were evaluated in a retrospective study; lab-to-lab variability was the greatest source of error. To this end, a number of NIH institutes and private-sector technology providers have established proficiency testing programs that utilize identical samples and amplification technologies to measure lab-to-lab variability.13 The latter comparison, which focuses on protocol comparability, yielded intriguing results. As suspected, technologies from the same manufacturer (in this instance Affymetrix one-round protocol and two-round protocol) exhibited the highest level of correlation at r2 = 0.96. Enzo’s protocol, which utilizes the same RT reagents on the front end of the protocol as Affymetrix, had the next best correlation at r2 = 0.94. This was followed by Ambion’s two-round protocol at r2 = 0.89, which utilizes the same T7 amplification approach with completely independent reagents. Lastly, NuGEN’s correlation was at r2 = 0.84, which one might expect given the RT reagents and nucleic acid used for target hybridization (cDNA) is completely different than in the T7 approach. Although the consensus is that data are comparable across the best-performing two-round protocols, clearly the most easy comparisons from a computational perspective (when compared to one-round T7 protocol) are within the Affymetrix product line. It is also important to keep in mind that the manner with which comparability is measured is insensitive to the magnitude and functional content of the information being examined. To this end, more detailed functional analyses with respect to probe specificity, reproducibility, and biological interpretation were performed and discussed in this study.
The recommendations for implementing small-sample amplification approaches into a laboratory workflow are as follows:
Learn the protocols
Take the time to learn the theory and technical minutia for small-sample amplification protocols. Each vendor has optimized the reagent composition and process to ensure reproducible performance. For many protocols, we found that formal training by the vendor led to significantly improved performance and that all information is not easy to convey in a technology manual. With appropriate training (and expertise), all small-sample amplification protocols can be performed at a high level of proficiency. With that said, the protocols that require fewer manipulations reduce time to arrays and reduce the potential for error when working with large sample numbers.
Know the stopping points
For protocols that require multiple days of procedures (and even the ones that don’t) the experimentalist should be very aware of the safe places to stop and store interim materials. Once again, the manufacturers have benchmarked these stop gaps, and users can help ensure success by stopping and storing biologicals at an appropriate time in the protocol. Some steps, although theoretically a good choice, are often not, in light of downstream processes.
Establish good performance metrics
Irrespective of the platforms tested and analyzed, this study has demonstrated the importance of identifying metrics that can be used to judge the quality of amplification technologies. The approaches described herein for assessing probe-level performance irrespective of platform has implications beyond the scope of this study. Furthermore, understanding the relative contribution of probe sets for a given gene annotation is important for many other aspects of data interpretation. Certainly the approach that has the highest correlation of probes that behave in a similar manner, as is seen in some of the protocols reviewed, is of paramount importance for downstream data interpretation.
Importance of target validation
The importance of target (i.e., differentially expressed genes) validation is essential. To validate whether or not the array is reporting accurate changes in gene expression, most laboratories have adopted quantitative real-time PCR to provide this information. Given that there is a subset of differentially expressed genes that differ between amplification technologies, it was important that some of these be tested to better understand the merits of one technology over another. To this end, given that the biggest differences in differentially expressed genes were between NuGEN and 2X T7 (with NuGEN providing a much longer list of differentially expressed genes not detected by T7), a list of targets was selected from this comparison. All genes tested by QPCR validated microarray results. In other words, all of the novel genes identified by NuGEN and not detected by T7 behaved in the same manner as the array reports. The inverse was also true, leading us to the conclusion that one approach is not necessarily better than the other just different. In fact, the approaches play nicely off each other in that they provide additional information to an already rich data set. However, it is important to note that the NuGEN protocol identifies significantly more genes than T7, and although all of the genes were validated in this study, it does not ensure that all will behave accordingly. Given the increased number of differentially expressed genes in the NuGEN protocol, more potential leads are likely.
In summary, all small-sample amplification technologies are not created equal, but all perform within the published specifications provided by the manufacturer. In addition, several metrics and analytical tools were identified that can be used irrespective of amplification approach in order to judge sample quality and sample-to-sample reproducibility. Throughout this process, the study has yielded many interesting conclusions that can be used to help laboratories performing nucleic acid amplification achieve the most reproducible and sensitive data sets. Irrespective of amplification protocols, the transition to the use of small samples from limiting RNA sources is an experimental reality. Clearly, given the differences between a single round of amplification and any of the small-sample amplification approaches investigated, the reproducibility within a given protocol is higher than across protocols. With that said, investigators must make good choices when deciding on an amplification approach for their studies. If the experimentalist uses a nanogram quantity of RNA (even though there may be more for some samples), this helps ensure that the data generated will still be useable when the project becomes more refined (i.e., when the investigator moves from tissue punches to FACS sorted samples or laser capture microdissection, or when a biopsy yields much less RNA, etc.). Another important conclusion revolves around the issue of historical data. For a laboratory that has a large amount of single-round amplification data and wants to have the best chance of comparing data at the raw signal level, it is best to continue with multiple rounds of the same amplification technology. This is not because it may be more sensitive or specific, but rather more closely aligned due to the use of enzymes and processes that can be more directly correlated to the original data set. Once again, although the biological output may be similar, those who want to compare data at the most basic level (i.e., signal value) should be cognizant of choosing the appropriate technology. For the laboratory starting to utilize this technology or for those programs that are experimenting with small samples for the first time, the choice of amplification approaches can be made using the information provided in this study. Using a protocol that is less labor intensive, applying a technology that has good reproducibility from sample sample, and employing an approach that maximizes sensitivity and specificity are of paramount importance.
All small-sample amplification approaches performed per the manufacturer’s claims; however, a difference in data quality does exist across protocols. The Affymetrix two-round protocol performed the best when compared to the one-round Affymetrix protocol, as one might expect, given that the same enzymes and protocols used the tighter correlation between one- and two-round (on a raw data level) data. However, NuGEN’s Ovation protocol (which yields cDNA instead of cRNA/aRNA) provided the most sensitive and specific data among the small-sample amplification protocols (yielding the most differentially expressed genes). With that said, the correlation between NuGEN and one round of Affymetrix labeling was the lowest (due to the fundamental differences in hybridization of DNA and RNA, or possibly the difference in sensitivity and/or specificity leading to a larger number of differentially expressed genes). Even approaches that had the biggest difference with one round of T7 demonstrated a good correlation of differential gene expression, truly making any of these technologies a viable option for laboratories with existing data or those who are just getting started.
ACKNOWLEDGMENTS
This research was supported in part by the NIEHS sponsored UMDNJ Center for Environmental Exposures and Disease, Grant #NIEHS P30ES005022.
REFERENCES
- 1.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, et al. Molecular classification of cancer: Class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 2.Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 3.Ma X-J, Salunga R, Tuggle JT, Gaudet J, Enright E, McQuary P, et al. Gene expression profiles of human breast cancer progression. Proc Natl Acad Sci USA. 2003;100:5974–5979. doi: 10.1073/pnas.0931261100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Affymetrix GeneChip Expression Analysis Technical Manual (2001, rev. 2004) Affymetrix, Inc; Santa Clara, CA: [Google Scholar]
- 5.Lipshutz RJ, Fodor SP, Gingeras TR, Lockhart DJ. High density synthetic oligonucleotide arrays. Nat Genet. 1999;21S:20–24. doi: 10.1038/4447. [DOI] [PubMed] [Google Scholar]
- 6.Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zuang Z, Goldstein SR, et al. Laser capture microdissection. Science. 1996;27:998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- 7.Sugiyama Y, Sugiyama K, Hirai Y, Akiyama F, Hasumi K. Microdissection is essential for gene expression profiling of clinically resected cancer tissues. Am J Clin Pathol. 2002;117:109–116. doi: 10.1309/G1C8-39MF-99UF-GT2K. [DOI] [PubMed] [Google Scholar]
- 8.van Gelder RN, von Zastrow ME, Yool A, Dement WC, Barchas JD, Eberwine JH. Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc Natl Acad Sci USA. 1990;87:1663–1667. doi: 10.1073/pnas.87.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phillips J, Eberwine JH. Antisense RNA amplification: A linear amplification method for analyzing the mRNA population from single living cells. Methods. 1996;10:283–288. doi: 10.1006/meth.1996.0104. [DOI] [PubMed] [Google Scholar]
- 10.Mahadevappa M, Warrington JA. A high-density probe array sample preparation method using 10- to 100-fold fewer cells. Nat Biotechnol. 1999;17:1134–1136. doi: 10.1038/15124. [DOI] [PubMed] [Google Scholar]
- 11.Dafforn A, Chen P, Deng G, Merrler M, Iglehart D, Koritala S, et al. Linear mRNA amplification from as little as 5 ng total RNA for global gene expression analysis. BioTechniques. 2004;37:854–857. doi: 10.2144/04375PF01. [DOI] [PubMed] [Google Scholar]
- 12.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 13.Brooks AI, Viale A. Assessing, understanding and minimizing variability of DNA microarrays. Am Pharm Rev. 2003;6(4):102–105. [Google Scholar]