Abstract
Comprehensive proteomic analyses require efficient and selective pre-fractionation to facilitate analysis of post-translationally modified peptides and proteins, and automated analysis workflows enabling the detection, identification, and structural characterization of the corresponding peptide modifications. Human serum contains a high number of glycoproteins, comprising several orders of magnitude in concentration. Thereby, isolation and subsequent identification of low-abundant glycoproteins from serum is a challenging task. selective capturing of glycopeptides and -proteins was attained by means of magnetic particles specifically functionalized with lectins or boronic acids that bind to various structural motifs. Human serum was incubated with differentially functionalized magnetic micro-particles (lectins or boronic acids), and isolated proteins were digested with trypsin. Subsequently, the resulting complex mixture of peptides and glycopeptides was subjected to LC-MALDI analysis and database searching. In parallel, a second magnetic bead capturing was performed on the peptide level to separate and analyze by LC-MALDI intact glycopeptides, both peptide sequence and glycan structure. Detection of glycopeptides was achieved by means of a software algorithm that allows extraction and characterization of potential glycopeptide candidates from large LC-MALDI-MS/MS data sets, based on N-glycopeptide-specific fragmentation patterns and characteristic fragment mass peaks, respectively. By means of fast and simple glycospecific capturing applied in conjunction with extensive LC-MALDI-MS/MS analysis and novel data analysis tools, a high number of low-abundant proteins were identified, comprising known or predicted glycosylation sites. According to the specific binding preferences of the different types of beads, complementary results were obtained from the experiments using either magnetic ConA-, LCA-, WGA-, and boronic acid beads, respectively.
Keywords: Glycosylation, Concanavalin A, Lens culinaris agglutinin, wheat germ agglutinin, boronic acid, magnetic particles, LC-MALDI
Protein glycosylation is the most common post-translational modification and plays a key role in protein stability, solubility, folding, and activity. Further, cellular processes such as protein trafficking and sorting and molecular recognition are dependent on glycosylation.1 Thus, glycosylated proteins comprise an interesting sub-proteome, which will have to be closely explored by in-depth proteomic analyses to obtain a better understanding of many biological processes. Additionally, it has been shown that aberrant, additional, or missing glycosylation is correlated with certain diseases, including cancer or congenital disorders of glycosylation (CDG).2–5
Traditional proteomics techniques based on 2D gel electrophoresis methods are very time consuming and laborious. The identification of glycosylated proteins using this strategy is often difficult due to the heterogeneity of the glycan structures, resulting in a distribution of a respective protein over different gel spots. Further, the work with samples like serum or plasma is hampered by the composition of the sample itself, which consists of estimated tens of thousands of proteins spanning a dynamic range of 10 orders of magnitude.6 The distribution of these proteins is dominated by a few highly abundant proteins, accounting for 99% of the total proteome of serum.7 Therefore, the specific isolation of glycosylated species is an important prerequisite for advanced analysis of glycoproteins and -peptides from complex biological sources. Different approaches for the specific isolation of glycosylated peptides and proteins have been previously described;8–10 e.g., lectin affinity chromatography employing different lectins, or covalent interaction chromatography with boronic acids or hydrazide chemistry. Lectins are proteins specifically recognizing and binding to defined glycostructures. Concanavalin A (ConA) is a lectin that binds mannosyl and glucosyl residues containing unmodified hydroxyl groups at positions C3, C4, and C611,12 and can be utilized for the targeted binding of certain oligosaccharide structures of N-glycosylated proteins.13 These binding requirements are fulfilled by the integral part of the conserved pentacore structure of N-linked oligosaccharides of the high-mannose type, the hybrid type, and the bi- and tri-antennary complex type. Lens culinaris agglutinin (LCA) is suitable for a more selective enrichment and recognizes hybrid or bi- and tri-antennary complex structures that are core fucosylated.14 The use of wheat germ agglutinin (WGA) allows for the isolation of glycostructures comprising N-acetylglucosamine and sialic acids.15 In contrast, phenyl boronic acid covalently reacts with 1,2-cis-diol group–containing molecules, and no complex recognition motif consisting of several saccharides is necessary for the interaction.16 All glycostructures containing saccharides like mannose, galactose, or glucose can form the heterocyclic diester, which is stable under moderate alkaline conditions. Thereby, boronic acids additionally facilitate the capturing of the more heterogeneous group of O-linked oligosaccharides.
Here, we describe a combination of specific glycoprotein isolation from human serum and LC-MALDI analysis, which facilitated the in-depth investigation of the serum glycoproteome. Differently functionalized magnetic particles were employed for capturing on the protein level to enrich large sets of glycoproteins for identification. The applied approach allowed for the investigation of additional glycomic-relevant questions—e.g., the identification of N-glycosylation sites. Further, a software tool was employed for the automated detection of glycopeptides, supporting the structural characterization of N-glycosylated peptides.
MATERIALS AND METHODS
Isolation of Glycoproteins
Glycoproteins from the serum of healthy volunteers were isolated using magnetic glyco-capturing beads (Bruker Daltonik GmbH, Germany). ConA, LCA, WGA, or boronic acids functionalized particles were employed for the specific capturing according to the manufacturer’s recommendations. Briefly, 20 μL of serum were incubated with 20 μL of magnetic particles in the provided binding buffer. After washing, the bound proteins were eluted under acidic conditions and dried in a vacuum centrifuge. The pellets were re-dissolved in 25 mM ammonium hydrogen carbonate (pH 8.2) and digested with 1 μg trypsin (Promega GmbH, Germany) at 37°C overnight. The reaction was stopped by addition of 1% TFA. Each capturing experiment was performed three times to provide enough material for the different paths of the work-flow (see Figure 1).
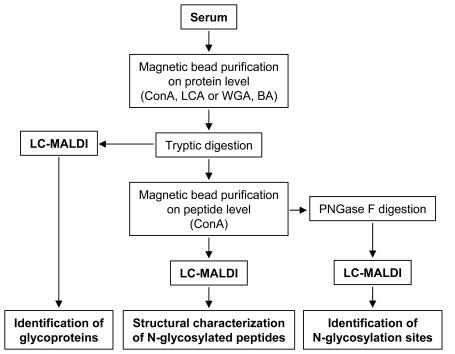
FIGURE 1.
Workflow describing the different paths for the identification of glycoproteins, the identification of N-glycosylation sites, and the structural characterization of N-glycosylated peptides from complex samples (human serum), respectively. (BA: boronic acids).
Identification of Glycoproteins
For the identification of serum glycoproteins, one part of the tryptic peptide digest obtained from the bead eluate was directly subjected to LC-MALDI analysis. LC separation was carried out using the following instrumental setup: LC system—Agilent 1100 capillary LC (Agilent Technologies, USA); column—C18 Pepmap 100 (180 μm I.D. × 15 cm, 5 μm, 100 A; LC Packings, The Netherlands); flow rate—2 μL/min; LC gradient—Eluent A, 0.1% aqueous TFA; Eluent B, 0.1% TFA in ACN. Gradient conditions: isocratic pre-run at 2% B, 0–5 min; linear gradient 2–40% B, 5–101 min; column wash at 80% B, 101–110 min; column conditioning at 2% B, 110–120 min. Online MALDI spotting of the LC fractions was carried as follows: spotting device, Proteineer FC (Bruker Daltonik, Germany); number of fractions, 384 (covering the gradient 2–40% eluent B); MALDI target, pre-spotted disposable anchorchip target PAC384 HCCA (Bruker Daltonik, Germany); fraction width, 15 sec (500 nL).
Isolation of Glycopeptides
Two residual parts of the trypsin-digested captured glycoproteins were dried in a vacuum centrifuge to remove ammonium hydrogen carbonate and TFA. The tryptic peptides were re-dissolved in 50 μL ConA wash buffer 2 of the kit and incubated with 20 μL ConA beads. After binding, the beads were washed three times with wash buffer 2 and the enriched glycopeptides were eluted under acidic conditions.
Identification of N-glycosylation Sites
To identify N-glycosylation sites, one part of the eluates containing the glycopeptides was dried in a vacuum centrifuge and the pellets were re-dissolved in 50 μL 25 mM sodium phosphate buffer (pH 7.5). N-glycans were released with 0.4 U PNGase F (Roche, Germany) at 37°C overnight. After drying in a vacuum centrifuge, the samples were re-dissolved in 10 μL 0.1% TFA and subjected to LC-MALDI analyses using the same conditions as for the identification of glycoproteins.
Structural Analysis of Intact Glycopeptides
For the characterization of intact glycopeptides, the remaining part of the enriched glycopeptides was directly subjected to LC-MALDI analysis using identical LC conditions as given above, with the exception of a steeper gradient from 2 to 50% eluent B. LC fractions were spotted on 600-μm anchorchip targets (Bruker Daltonik, Germany) using 2,5-DHB (1 μL per spot; 5 mg/mL dissolved in a mixture of 30% of 0.1% TFA, 70% ACN).
Mass Spectrometry
MS spectra were automatically acquired on an ultraflex III TOF/TOF (Bruker Daltonik GmbH, Germany) in the positive reflecton mode under the control of Compass 1.2 and WarpLC 1.1 software (Bruker Daltonik, GmbH, Germany). Detected peptide compounds with a signal-to-noise ratio higher than 10 were subjected to MALDI-TOF MS/MS.
Data Evaluation
Fragment ion spectra were subjected to database search employing MASCOT (www.matriscience.com) and NCBI sequence database (NCBInr 20051231—3149313 sequences, 1083633969 residues) restricted to Homo sapiens (human; 139753 sequences). The following search settings were applied: Type of search: MS/MS Ion Search; Enzyme: Trypsin; Variable modifications: Acetyl (N-term), Carbamidomethyl (C), Oxidation (M), Pyro-glu (N-term E), Pyro-glu (N-term Q); Mass values: Monoisotopic; Protein Mass: Unrestricted; Peptide Mass Tolerance: ± 100 ppm; Fragment Mass Tolerance: ± 0.7 Da; Max Missed Cleavages: 2; Instrument type: MALDI-TOF-TOF. Only peptides with p < 0.05 were considered significant hits. For identification of N-glycosylation sites, the variable modifications were extended to deamination of asparagine (N).
Fragment ion spectra of glycopeptides were first analyzed with FlexAnalysis 3.0 (Bruker Daltonik GmbH, Bremen, Germany) employing the integrated tool for glycopeptide analysis, which is described in detail later. The identified mass of the peptide part ([M+H]+) was subsequently used as parent mass for the MASCOT search employing BioTools 3.1 software (Bruker Daltonik GmbH, Bremen, Germany).
RESULTS AND DISCUSSION
The aim of the workflow described here was to cover various aspects of glycoproteomics, and thus, by means of sophisticated glycospecific sample pre-treatment, to identify serum glycoproteins, to detect N-glycosylation sites, and finally to characterize intact glycopeptides—both their peptide sequence and glycan structure—from MS/MS data. The complete workflow is depicted in Figure 1.
Identification of Glycoproteins
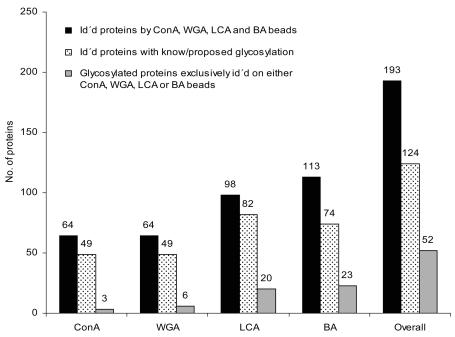
The results of the LC-MALDI analyses of the isolated glycoproteins revealed an overall identification of 193 proteins comprising 124 proteins with known or proposed glycosylation sites (Figure 2). The apparently high number of nonspecifically isolated proteins includes a number of nonglycosylated proteins that are known to be associated with glycosylated ones. As an example, the nonglycosylated complement C8 β chain was identified, which forms a complex with the glycosylated complement C8α. Fifty-two of the glycosylated proteins were exclusively isolated by ConA, WGA, LCA, or boronic acid beads, respectively. Each bead type represented its own individual binding profile, partially overlapping with the binding profiles of the other beads. The highest number of uniquely captured proteins was obtained from boronic acid beads. This is in accordance with the less restrictive binding of the boronic acid beads, requiring only vincinal cis-diol groups to form the heterocyclic boronic diester between boronic acids and the saccharide. The selectivity of the applied capturing approach was demonstrated by the identification of the medium-abundant serum glycoprotein properdin (0.01 g/L serum), which was found only within the boronic acid fraction. According to the SwissProt database (www.expasy.org), this protein comprises 14 C-, one O-, and one N-glycosylation site. C-glycosylation consists of a single mannosyl residue linked via the C2 of the indole ring of tryptophan, and represents the ideal binding motif for boronic acids. Thereby, it is most likely that properdin had been captured via its C-glycosylation sites.
FIGURE 2.
Summary of the numbers of identified proteins and glycoproteins after isolation by the different bead types. Total numbers of identified proteins, black bars; numbers of identified glycoproteins, dotted bars; numbers of glycoproteins exclusively identified by a single bead type, gray bars.
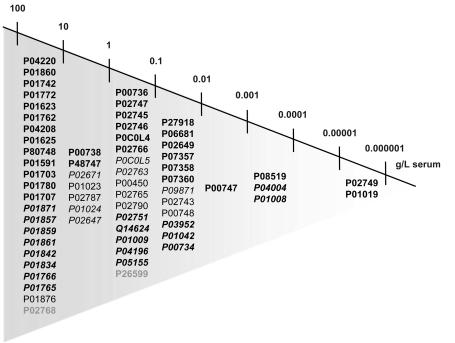
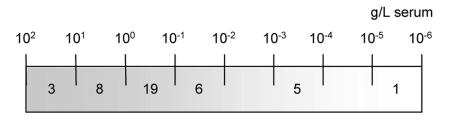
The sensitivity of the magnetic particles facilitated the capturing of serum glycoproteins over a large concentration range, spanning eight orders of magnitude (Figure 3). One of the lowest-abundant proteins identified was β-2-glycoprotein, which is present in serum at a concentration of 1 × 10−6 g/L. The specificity of the glyco-capturing beads was demonstrated by the finding that none of the high-abundant serum proteins was isolated by all four different bead types. In case of insufficient specificity, the binding of high-abundant proteins would have to be expected for all bead types.
FIGURE 3.
Dynamic range of identified glycoproteins isolated by the different bead types, spanning eight orders of magnitude. Proteins are listed according the SwissProt accession numbers, so their serum concentration was known. Proteins identified by only one bead type (bold), proteins identified by two different bead types (italic), proteins identified by three bead types (normal), proteins identified by all four bead types (italic and bold), and nonglycosylated, identified proteins (gray).
The detection and identification of glycoproteins is an important issue in glycomics. To obtain more detailed information about the glycoproteins, further investigations are necessary. An important question in glycomic analysis is the identification of N-glycosylation sites. Both, the number and the position(s) of glycosylation are of interest.
Identification of N-glycosylation Sites
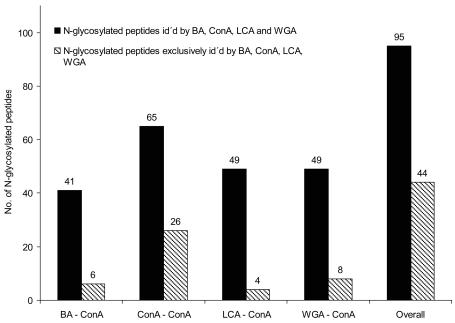
The identification of N-glycosylation sites required a modification and an extension of the workflow described above (Figure 1). The complexity of the peptide mixture after the tryptic digestion was reduced by second magnetic bead purification on the peptide level, exemplarily employing ConA beads. The enriched glycopeptides were deglycosylated with PNGase F and subsequently subjected to LC-MALDI analysis. In total, 95 N-glycosylation sites were identified (Figure 4), representing the ConA-affine fraction of glycopeptides in serum. Most serum proteins contain highly complex glycans, which only to a certain extent comprise the required binding motif for ConA. Due to the specificity of ConA, only nonfucosylated, bi-, and tri-antennary glycostructures were isolated, representing a small subset of glycostructures present in serum. Accordingly, most glycopeptides were detected within the fraction captured by ConA on a protein and peptide level (ConA-ConA). This fraction also represented the most individual capturing profile. To get a more complete overview about the N-glycosylation sites in serum, further capturing experiments employing different combinations of bead types would be necessary. The applied approach facilitated the identification of even very low-abundant N-glycosylation sites. Figure 5 depicts the distribution of identified sites with respect to the original protein concentration in serum. N-glycosylation site N162 of β-2-glycoprotein with a serum concentration of 1 × 10−6 g/L serum was the site with the lowest concentration that had been identified.
FIGURE 4.
Numbers of N-glycosylation sites identified after first bead purification on the protein level employing different bead types, and second bead purification using ConA. Total numbers of identified sites, black bars; numbers of N-glycosylation sites exclusively identified by a single bead type, hatched bars.
FIGURE 5.
Numbers of identified N-glycosylation sites in correlation to the serum concentration of the corresponding glycoprotein as far as the concentration was known.
The digestion with PNGase F is a well established approach to verify predicted N-glycosylation sites or to identify new ones. In certain cases, not only the presence and position of the glycosylation but the glycan structure at a particular position is of interest. For example, aberrant glycosylations, like additional or missing fucosylations, are known to be correlated to diseases.2 The approach described above does not provide any information about the glycan part, and even the analysis of the removed glycans does not provide this information, because the connectivity between peptide and glycan has been lost during PNGase F digest. This problem can be circumvented by direct analysis of intact N-glycosylated peptides.
Structural Analysis of Intact Glycopeptides
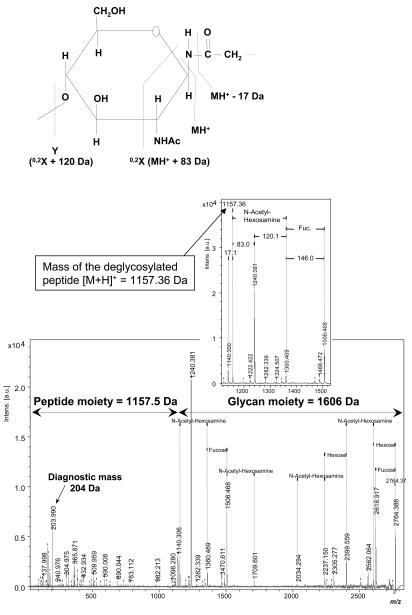
The fragmentation of N-glycosylated peptides results in characteristic peak patterns in MALDI-MS/MS spectra, which can yield structural information about the peptide and glycan parts from the same spectrum. All N-glycosylated proteins comprise a conserved core structure consisting of two N-acetylglycosamine residues linked to asparagine followed by three branched mannosyl residues. During MS/MS fragmentation, the inner core N-acetylglucosamine undergoes a 0,2X cross-ring cleavage, resulting in mass differences of 120 and 83 Da (Figure 6a). A subsequent elimination of ammonia from the previously glycosylated asparagine leads to a further mass shift of 17 Da. This characteristic peak pattern of delta 17-83-120 Da is unique for the fragmentation of N-glycosylated peptides, allowing the identification of the [M+H]+ mass of the corresponding peptide without its glycan part (Figure 6b). Further, a single N-acetylglucosamine is detected as a diagnostic mass at about 204 Da. If the glycan core is fucosylated, the characteristic pattern of 17-83-120 Da is extended by 146 Da, corresponding to a single fucose. Sialic acids containing N-glycosylated peptides show mass shifts of 291 Da, corresponding to neutral losses of single sialic acid residues. Since sialylation is a very labile modification, only the number of observed sialic acids of the analyzed peptide can be determined, not the number of sialic acids in the original peptide. From the N-acetylglucosamine-related fragment pattern described above, the mass of the peptide moiety and glycan moiety can be derived. This allows running such an MS/MS spectrum in a MASCOT database search to identify the respective peptide sequences. Alternatively, for peptide sequence determination, the data set that was obtained from PNGase F digestion performed on the same sample in parallel can be screened for the corresponding peptide showing a 1-Da-higher mass (deamidation at N-glycosylation site induced by the deglycosylation enzyme). For the characterization of the glycan composition and structure from MALDI-MS/MS data of intact N-glycopeptides, FlexAnalysis software can be used to annotate in the spectra mass distances that are typical for common glycan structures. This at least allows deduction of the composition of the glycan regarding various types of sugar units (hexose, fucose, pentose, hexNac, sialic acid).
FIGURE 6.
A: Fragmentation scheme of the asparagin linked N-acetylglucosamine residue of the conserved pentacore of N-linked glycans. The 0,2X cross ring cleavage and subsequent elimination of ammonia result in characteristic mass differences of 17, 83 and 120 Da. The peak between the differences of 17 and 83 Da (1175 Da) corresponds to the mass of the peptide part ([M+H]+ of the deglycosylated peptide. B: MALDI-TOF MS/MS fragment ion spectrum of an N-glycosylated peptide after analysis with Flexanalysis and application of the detection tool for glycopeptides. After detection of the mass of the deglycosylated peptide the subsequent MASCOT search revealed a fragment of immunoglobulin γ chain.
To make these search parameters available for automated analysis, the search algorithm has been integrated as a script-based tool into the FlexAnalysis software, making it possible to analyze intact glycopeptides to obtain structural information about the peptide as well as the glycan moiety from the same MALDI-TOF MS/MS spectrum. Figure 6b shows an example of the fragment ion spectrum of an N-glycosylated peptide and its respective annotation after analysis with this software tool. The calculation of the mass of the glycan moiety permitted a suggestion about its potential glycan composition. The mass of the peptide part [M+H]+ was used as parent mass for a Mascot search to identify the corresponding peptide sequence. In this case, a glycopeptide of human immunoglobulin γ chain and its respective glycosylation has been identified. The mass of the glycan moiety corresponds to the composition HexNAc2FucHex3HexNAc2Hex, which is in concordance with the previously described complex-type glycosylation of immunoglobulin.17
CONCLUSIONS
The described workflow facilitates several parallel investigations important for glycomic analysis. Functionalized magnetic particles allow for the sensitive capture of glycoproteins from complex samples, with a detection range of identified glycoproteins spanning eight orders of magnitude. The use of differently functionalized particles for the glycoprotein isolation reveals individual but overlapping binding profiles for the different bead types. The sensitivity of the capturing procedure permits the identification of low-abundant N-glycosylation sites (1 × 10−6 g/L in serum).
LC-MALDI-MS/MS analysis using 2,5-DHB as a most suitable glycopeptide matrix applied in conjunction with software-driven detection of glycopeptide candidates has proven to be a very useful method to characterize intact glycopeptides regarding both the incorporated peptide and glycan structure. This approach yields valuable information about protein glycosylation, which cannot be derived from well established methods such as separate analysis of the peptide and glycan moiety after PNGase F treatment.
While the software script as described above is of great help in the extraction and characterization of potential glycopeptide candidates from extensive MS/MS data sets, the evaluation of the obtained result lists is still a challenge with respect to its complexity. However, for further typical glycoprotein analysis tasks, for instance, LC-MALDI-MS/MS analysis of isolated digested glycoproteins and analysis of digested lower-complexity glycoprotein mixtures, this approach works out very efficiently. Further optimization of the experimental setup will be required to allow a broader range of N-glycosylated peptides to be efficiently characterized from complex samples such as human serum.
ACKNOWLEDGMENT
This work was supported by the Sächsische Aufbaubank grant 7495/1185.
REFERENCES
- 1.Ohtsubo K, Marth JD. Glycosylation in cellular mechanisms of health and disease. Cell. 2006;126(5):855–867. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 2.Durand G, Seta N. Protein glycosylation and diseases: Blood and urinary oligosaccharides as markers for diagnosis and therapeutic monitoring. Clin Chem. 2000;46 (6):795–805. [PubMed] [Google Scholar]
- 3.Miyamoto S. Clinical applications of glycomic approaches for the detection of cancer and other diseases. Curr Opin Mol Ther. 2006;8(6):507–513. [PubMed] [Google Scholar]
- 4.Freeze HH, Aebi M. Altered glycan structures: The molecular basis of congenital disorders of glycosylation. Curr Opin Struct Biol. 2005;15(5):490–498. doi: 10.1016/j.sbi.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 5.Leroy JG. Congenital disorders of N-glycosylation including diseases associated with O- as well as N-glycosylation defects. Pediatr Res. 2006;60(6):643–656. doi: 10.1203/01.pdr.0000246802.57692.ea. [DOI] [PubMed] [Google Scholar]
- 6.Anderson NL, Anderson NG. The human plasma proteome: History, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1(11):845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 7.Chan KC, Lucas DA, Hise D, Schaefer CF, Xiao Z, Janini GM, et al. Analysis of the human serum proteome. Clin Proteomics. 2004;1:101–225. [Google Scholar]
- 8.Bunkenborg J, Pilch BJ, Podtelejnikov AV, Wiśniewski JR. Screening for N-glycosylated proteins by liquid chromatography mass spectrometry. Proteom. 2004;4:454–465. doi: 10.1002/pmic.200300556. [DOI] [PubMed] [Google Scholar]
- 9.Zhang H, Yi EC, Li X-j, Mallick P, Kelly-Spratt KS, Masselon CD, et al. High throughput quantitative analysis of serum proteins using glycopeptide capturing and liquid chromatography mass spectrometry. Mol Cell Proteomics. 2005;4(2):144–155. doi: 10.1074/mcp.M400090-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Yang Z, Hancock WS. Approach to the comprehensive analysis of glycoproteins isolated from human serum using multi-lectin affinity column. J Chromatogr A. 2004;1053(1–2):79–88. [PubMed] [Google Scholar]
- 11.Kamra A, Gupta MN. Crosslinked concanavalin A-O-(diethylaminoethyl)-cellulose—an affinity medium for concanavalin A-interacting glycoproteins. Anal Biochem. 1987;164 (2):405–410. doi: 10.1016/0003-2697(87)90511-2. [DOI] [PubMed] [Google Scholar]
- 12.Yahara I, Edelman GM. Restriction of the mobility of lymphocyte immunoglobulin receptors by Concanavalin A. PNAS. 1972;69 (3):608–612. doi: 10.1073/pnas.69.3.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldstein IJ, Hollerman CE, Smith EE. Protein-carbohydrate interaction. II. Inhibition studies on the interaction of Concanavalin A with polysaccharides. Biochemistry. 1965;41:876–883. doi: 10.1021/bi00881a013. [DOI] [PubMed] [Google Scholar]
- 14.Young NM, Leon MA, Takahashi T, Howard IK, Sage HJ. Studies on a phytohemagglutinin from the lentil. 3. Reaction of Lens culinaris hemagglutinin with polysaccharides, glycoproteins, and lymphocytes. J Biol Chem. 1971;246(6):1596–1601. [PubMed] [Google Scholar]
- 15.Nagata Y, Burger MM. Wheat germ agglutinin. Molecular characteristics and specificity for sugar binding. J Biol Chem. 1974;240:3116. [PubMed] [Google Scholar]
- 16.Rawn JD, Lienhard GE. The binding of boronic acids to chymotrypsin. Biochemistry. 1974;13 (15):3124–3130. doi: 10.1021/bi00712a019. [DOI] [PubMed] [Google Scholar]
- 17.Schuster M, Umana P, Ferrara C, Brunker P, Gerdes C, Waxenecker G, et al. Improved effector functions of a therapeutic monoclonal Lewis Y-specific antibody by glycoform engineering. Cancer Res. 2005;65(17):7934–41. doi: 10.1158/0008-5472.CAN-04-4212. [DOI] [PubMed] [Google Scholar]