Abstract
Cytokines and chemokines are responsible for regulating inflammation and the immune response. Cytokine and chemokine release is typically measured by quantitative enzyme-linked immunosorbant assay (ELISA) or Western blot analysis. To expedite the analysis of samples for multiple cytokines/chemokines, we have developed slide-based Thermo Scientific ExcelArray Antibody Sandwich Microarrays. Each slide consists of 16 subarrays (wells), each printed with 12 specific antibodies in triplicate and positive and negative control elements. This 16-well format allows for the analysis of 10 test samples using a six-point standard curve. The array architecture is based on the “sandwich” ELISA, in which an analyte protein is sandwiched between an immobilized capture antibody and a biotinylated detection antibody, using streptavidin-linked Thermo Scientific DyLight 649 Dye for quantitation. The observed sensitivity of this assay was <10 pg/mL. In our experiments, the Jurkat cell line was used as a model for human T-cell leukemia, and the A549 cell line was used as a model for human non–small cell lung cancer. To evoke a cytokine/chemokine response, cells were stimulated with tumor necrosis factor alpha (TNFα), phorbol-12-myristate-13-acetate (PMA, TPA), and phytohemagglutinin (PHA). Cell supernatants derived from both untreated and stimulated cells were analyzed on four different arrays (Inflammation I, Inflammation II, Angiogenesis, and Chemotaxis), enabling the quantitation of 41 unique analytes. Stimulated cells showed an increase in the expression level of many of the test analytes, including IL-8, TNF-α, and MIP-1α, compared to the non-treated controls. Our experiments clearly demonstrate the utility of antibody microarray analysis of cell-culture supernatants for the profiling of cellular inflammatory mediator release.
Keywords: protein microarrays, inflammation, multiplexing, ExcelArray
Cytokines are important molecules that play an integral role in the initiation and promotion of most immune responses. These proteins have important functions in the regulation of pathways governing inflammation, angiogenesis, and cellular chemotaxis. The ability to rapidly and accurately determine the effect various stimuli have upon cytokine levels is important to the research, clinical, and diagnostic communities. In support of these efforts, we have developed four human 12-plex cytokine/chemokine antibody microarrays featuring many improvements over currently available alternatives.
The microarray, or biochip, allows for the simultaneous screening of tens to tens of thousands of selected biological targets. As the use of microarray technology in genotyping and gene expression experiments has developed over the past decade, nucleic acid arrays are widely produced and utilized in numerous core facilities and research laboratories.1–3 Protein microarray technology is not as developed as nucleic acid microarray technology, but the potential uses of protein microarrays are equal to, if not greater than, those of the nucleic acid microarray. Examples of the utility of protein microarrays as screening tools for protein expression and protein interactions are abundant.4–9 The multiplex antibody microarray format offers several important advantages over the traditional single-plex enzyme-linked immunosorbant assay (ELISA), including time, cost, and sample/reagent savings.
Multiplexed antibody microarrays are protein microarrays that, when properly designed and validated, result in a high-throughput immunoassay platform capable of generating high-quality data with low-pg/mL sensitivity. To realize this level of sensitivity, antibody microarrays must display low background levels, minimal antibody cross-reactivity, and minimal spot size and volume variability. Although both assay types (ELISA and antibody microarray) typically use the same materials in assay construction (i.e., capture antibodies, analytes, detection antibodies, and a reporting system such as fluorescence, chemiluminesence, etc.), there are inherently more difficulties in validating the performance of a multiplexed antibody microarray compared to a single-plex ELISA. This increased difficulty is due to the requirement that the materials used to detect each specific analyte, when tested in combination with other analytes simultaneously, not significantly cross-react. Cross-reactivity, the nonspecific interaction of assay components with unintended targets, skews calculated analyte concentrations. Additionally, sample diluent and other assay reagents and formulations must be optimized to ensure simultaneous performance of all capture antibody-analyte-detector antibody combinations.
We have constructed four antibody microarrays for the detection of cytokines and chemokines released by cells during human inflammation, angiogenesis, and cellular chemotaxis processes (Table 1, Figure 1). These microarrays are printed on PATHplus Microarray Slides. The PATHplus surface allows for increased dynamic range, high signal-to-noise ratios, and good reproducibility compared with conventional derivatized glass and nitrocellulose pad slides. Streptavidin-conjugated DyLight 649, a fluorescent molecule exhibiting superior photostability and fluorescence intensity, is utilized as the detector molecule.
TABLE 1.
Microarray Content
| Thermo Scientific ExcelArray Inflammation I | Thermo Scientific ExcelArray Inflammation II | Thermo Scientific ExcelArray Angiogenesis | Thermo Scientific ExcelArray Chemotaxis |
|---|---|---|---|
| IFNγ | IL-1α | Angiogenin | Eotaxin |
| IL-1β | IL-3 | EGF | I-309 |
| IL-2 | IL-5 | FGFb | IP-10 |
| IL-4 | IL-7 | GROα | MCP-3 |
| IL-6 | IL-13 | HGF | MDC |
| IL-8 | IL-15 | IL-8 | IL-8 |
| IL-10 | IL-17 | HB-EGF | Fractalkine |
| IL-12p70 | FasL | KGF | TARC |
| RANTES | G-CSF | PlGF | RANTES |
| MIP-1α | GM-CSF | VEGF | MIP-1α |
| MIP-1β | TNFβ | TIMP-1 | MIP-1β |
| TNFα | MCP-1 | TNFα | MCP-1 |
The protein targets for the four human cytokine/chemokine arrays are tabulated.
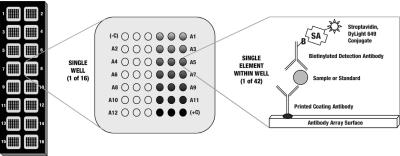
FIGURE 1.
Microarray layout. A1–A12 signifies the 12 analyte positions available; these positions correspond to the targets listed in Table 1. (−C) and (+C) indicate the positions of the negative and positive control elements, respectively.
All components of these antibody microarrays were developed to maximize performance in a multiplexed format. All assay standards are calibrated and validated against National Institute for Biological Standards and Controls (NIBSC) standards (when available) to ensure accurate analyte quantification. Finally, the performance of each assay was validated by analyzing both null samples spiked with recombinant protein standards and natural samples expressed from stimulated human peripheral blood mononuclear cells (PBMCs).
Presented here is work compiled during the development of these microarrays and applications work in which the cytokine/chemokine response of the Jurkat human T-cell leukemia cell line and the A549 human non–small cell lung carcinoma cell line were monitored. Cell culture supernatant samples were assayed on the four antibody microarrays, allowing for the simultaneous analysis of 41 different cytokine/chemokine analytes.
MATERIAL AND METHODS
Microarray Production
Each of the four microarray panels was printed onto PATHplus Thin-film Nitrocellulose Slides (GenTel BioSciences, Madison WI). The concentration of each capture antibody was titered to maximize signal generation when antibodies are diluted in a proprietary print buffer. This print buffer provides optimal spot morphology and spot volume consistency. Each capture antibody was printed in triplicate, at a volume of 2 nL and nominal spot diameter of 250 μm, into 16 subarrays in a 2 column by 8 row format. Each subarray also contained triplicate positive and negative control antibody elements. After the slides were printed and checked for element quality and consistency, they were treated according to a custom-designed process that preserves spot morphology and spot-to-spot consistency. Briefly, the printed slides were blocked using a proprietary buffer formulation and process predetermined to minimize nonspecific adsorption, preserve signal strength, and greatly enhance antibody stability. The blocked slides were then dried and fitted with silicone-backed, reaction well–defining overlays (16 wells, one for each subarray). The completed slide assembly was then vacuum sealed to maximize antibody and preprinted slide stability.
Validation of Antibody Cross-Reactivity
The capture and detector antibodies, as well as the recombinant antigen standards, were analyzed for cross-reactivity as follows. Preprinted slides were assayed and each sub-array was probed with a single antigen, rinsed and incubated with biotinylated detection antibodies, rinsed again, and finally probed with streptavidin-labeled fluorophore. Signal intensity was measured across all features to determine capture antibody–antigen cross-reactivity. It is important to note that the zero-concentration analyte well of the standard curve used for quantitation also provided control against signal generated by the nonspecific interaction of detector antibodies. Our criterion for acceptable cross-reactivity was that antibody pairs should not cross-react with any target at a level greater than 5% of the signal generated from the specific interaction of the individual analyte.
Spike and Recovery
Known amounts of each target analyte were spiked into human serum and cell-culture supernatants. These spiked samples were run on each of the four multiplexed arrays, and the percent recovery for each antigen was calculated according to the following equation:
The criterion for acceptable spike and recovery data is a recovery value between 80% and 120% of spiked amount.
Sensitivity Calculations
Microarray sensitivity was calculated for each analyte according to the following equation:
Desired sensitivity is less than 10 pg/mL for all analytes.
Natural Sample Recognition
To validate the ability of each multiplexed array to recognize naturally expressed targets, peripheral blood mono-nuclear cells (PBMCs) were stimulated as described below and the cell-culture supernatants were collected and analyzed on each of the four microarrays. The PBMCs were isolated using Ficoll-Hypaque density-gradient centrifugation of venous blood collected from a normal healthy donor and resuspended at 1 × 106 cells/mL in complete medium (RPMI + 10% fetal bovine serum [FBS] + 1X penicillin-streptomycin [Thermo Fisher Scientific (HyClone)]). To induce expression of as many targets as possible, PBMCs to be analyzed for each multiplexed array were stimulated by supplementing medium with 5 μg/mL PHA (phytohemag-glutinin [Inflammation I, Inflammation II, Angiogenesis and Chemotaxis], Sigma-Aldrich), 5μg/mL of concanavalin A (Inflammation I, Inflammation II and Angiogenesis, Sigma-Aldrich) or 5 ng/mL phorbol 12-myristate 13-acetate (PMA, Sigma-Aldrich) plus 500 ng/mL ionomycin (Inflammation II, Sigma-Aldrich). Finally, the PBMCs to be analyzed using the Chemotaxis array were stimulated with 1 μg/mL staphylococcal enterotoxin B fragment (SEB, Sigma-Aldrich). Supernatants were harvested after 2 d of incubation in the appropriately supplemented media and analyzed on the arrays. Nonstimulated control PBMC supernatants were also processed as a negative control for each multiplexed array.
Stimulation and Analysis of Jurkat Human T-Cell Leukemia Cells and A549 Human Non–Small Cell Lung Carcinoma Cells
Jurkat human T-cell leukemia cells and A549 human non–small cell lung carcinoma cells were obtained from ATCC (TIB-152 and CCL-185, respectively). These cells were cultured according to ATCC recommendations. Cytokine release in Jurkat cells was stimulated by addition of 7.5 nM PMA and 5 μg/mL PHA to media. Cytokine release in A549 cells was stimulated by addition of TNFα (Thermo Fisher Scientific) to medium at a final concentration of 25 ng/mL. Appropriate vehicle controls were performed for each cell treatment. Cell-culture supernatants were collected at 24–48 h and either assayed immediately or stored short term at –20°C.
Sample Analysis
Jurkat and A549 cell-culture supernatants prepared above were assayed for cytokine/chemokine concentration using Thermo Scientific ExcelArray Inflammation I (Product No. 82002), Inflammation II (No. 82003), Angiogenesis (No. 82004), and Chemotaxis (No. 82006) microarrays (Jurkat supernatants were not assayed on the Angiogenesis microarray). Assays were performed according to product instructions. Briefly, 100 μL of multiplexed standards (1000–12.3 pg/mL) or sample was applied to each well and incubated at room temperature for 2 h. After sample incubation, the microarrays were rinsed with wash buffer. Pre-titered biotinylated detector antibodies were applied to each microarray and incubated at room temperature for 1 h. Microarrays were rinsed and streptavidin-linked DyLight 649 was applied to the microarray and incubated for 30 min. The microarray was rinsed and dipped briefly in a final rinse solution. Microarrays were imaged using an AlphaScan Microarray Imager (Alpha Innotech), and spot densitometry was performed using ArrayVision software (GE Healthcare). The spot densitometry data generated were imported into Microsoft Excel software to calculate sensitivities, discrimination, and coefficients of variation (CVs).
RESULTS AND DISCUSSION
Traditionally, ELISA has been the method of choice for the quantification of cytokine and chemokine expression levels in cell-culture supernatants and serum samples. The antibody microarrays we developed and tested in this work were envisaged as a viable alternative to traditional ELISA. To be considered as such, the microarrays must meet the high performance standards exhibited by ELISA assays. The aim of this work was to validate the performance of our antibody arrays with respect to the following parameters: cross-reactivity, spike and recovery, sensitivity, and natural sample response.
Cross-reactivity results from nonspecific interaction of assay components with unintended targets. Cross-reactivity values for all arrays averaged less than 2%, with no one target exceeding a cross-reactivity value of 5% (Table 2). Minimal cross-reactivity is important for avoiding false positives. A cross-reactivity of less than 5% is acceptable for the desired level of assay performance. Cross-reactivity is a function of analyte homology and nonspecific antibody interactions. The antibody pairs that yielded the greatest levels of cross-reactivity recognized members of the same protein family, namely MIP-1α and MIP-1β, and MCP-1 and MCP-3.
TABLE 2.
Microarray Performance Testing Results
| Tested Parameter | ExcelArray Inflammation I | ExcelArray Inflammation II | ExcelArray Angiogenesis | ExcelArray Chemotaxis |
|---|---|---|---|---|
| Sensitivity (pg/mL; average, range) | 2.8, 0.8–6.5 | 3.4, 1.7–6.3 | 5.1, 1.5–9.3 | 1.25, 0.1–5.8 |
| Cross-reactivity of antibody pairs (%) | MCP-1&3 ≤5%
All others ≤1% |
All ≤4% | All ≤5% | All ≤2% |
| Recovery in Cell Culture Media (%;average, range) | 97%, 84–117% | 92%, 82–112% | 99.8%, 82–119% | 99.2, 86.9– 108.9% |
| Natural Sample Recognition (Yes/No, Stimulant) | Yes, PHA/Con A | Yes, PHA/Con A/PMA + Ionomycin | Yes, PHA/Con A | Yes, PMA/PHA |
| Standard Curve Accuracy | R2 ≥ 0.990 | R2 ≥ 0.999 | R2 ≥ 0.98 | R2 ≥ 0.98 (MCP-3) |
| Four parameter curve fit (average R2 value) | R2 = 0.990 (others) |
As outlined in Materials and Methods, the sensitivity, cross-reactivity, percent recovery, natural sample recognition, and standard curve accuracy were recorded for each analyte for each array.
Spike and recovery experiments are an important step in demonstrating the ability of an immunoassay to recognize sample analytes irrespective of sample matrix. This parameter is assessed as the ability to recover known amounts of recombinant proteins spiked into cell-culture supernatants. As an average percentage of spiked standard recovered, Inflammation I recovered 97% of spiked standards (range 84–117%), Inflammation II recovered 92% of spiked standards (range 82–112%), Angiogenesis recovered on average 99.8% of spiked standards (range 82–119%), and the Chemotaxis microarray averaged 99.2% recovery (range 86.9–108.9%) (Table 2). These are acceptable results for ELISA performance and represent excellent performance for a multiplex assay.
The determination of assay sensitivity is an important step in delineating the conditions under which an assay may be utilized. Sensitivity dictates the lower limit of analyte concentrations that may be reliably investigated and analyte concentration values assigned. Assay sensitivity values calculated for the Inflammation I, Inflammation II, Angiogensis, and Chemotaxis arrays were 2.8pg/mL (0.8–6.5 pg/mL), 3.4 pg/mL (1.7–6.3 pg/mL, 5.1 pg/mL (1.5–9.3 pg/mL), and 1.25 pg/mL (0.1–5.8 pg/mL), respectively (Table 2). Detection limits for all analytes met the sensitivity specifications, with several surpassing the desired sensitivity level (10 pg/mL) by more than one order of magnitude. The sensitivity value set for these assays was derived from commercially available colorimetric ELISA kit sensitivity specifications.
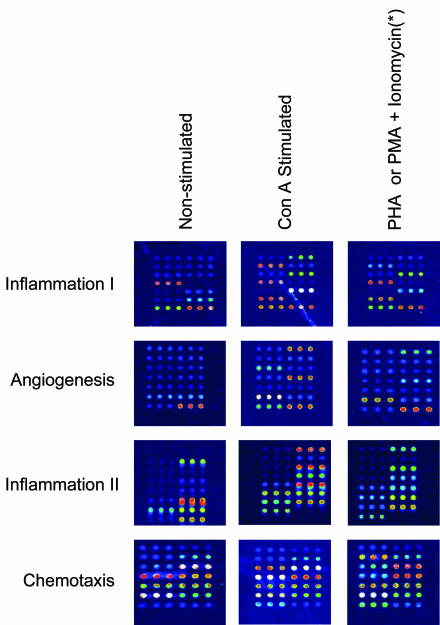
The detection of naturally expressed protein, as compared to recombinant protein, is an important facet of immunoassay performance. All four microarrays detected naturally occurring analyte proteins expressed in peripheral blood mononuclear cells (PBMC) (Table 2 , Figure 2). The detection of natural samples is important in that recombinant proteins often display antigenic epitopes differently than naturally produced proteins. By analyzing natural sample response, the validity of data generated from these microarrays is verified.
FIGURE 2.
Microarray false-color images of tested cell-cultured supernatants obtained from nonstimulated and stimulated peripheral blood mononuclear cells. (*) PHA used for Inflammation I, II, and Angiogenesis; PMA and ionomycin used for Chemotaxis.
Jurkat Human T-Cell Leukemia Cells and A549 Human Non–Small Cell Lung Carcinoma Cell Stimulation
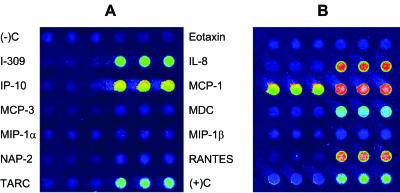
Two cell-culture model systems were employed to examine the ability of the antibody microarrays to accurately quantify cytokine levels in a multiplexed format. PHA/PMA stimulation of Jurkat human T-cell leukemia cells has been previously shown to induce release of various cytokines, including many contained on our four arrays.10 Treatment of A549 human non–small cell lung carcinoma cells with TNFα has similarly been shown to elicit a cytokine response.11,12 In our experiments, PHA/PMA stimulation of Jurkat cells results in increased expression levels of 14 cytokines and chemokines (14/31 targets analyzed) as compared to nonstimulated supernatants (Table 3). TNFα stimulation of A549 cells results in the increased expression of 12 cytokines and chemokines (12/41 targets analyzed) as compared to nonstimulated supernatants (Table 3). Interestingly, a significant decrease in the level of angiogenin was observed upon TNFα stimulation. Images of the unstimulated and TNFα-stimulated A549 cell-culture supernatants assayed on the Chemotaxis microarray reveal an increased protein expression level of seven target proteins (IL-8, I-309, IP-10, MCP-1, MDC, RANTES, and TARC) (Figure 3). These data are consistent with previously published data, specifically the increased expression of IL-8 as a result of TNFα stimulation.11,12
TABLE 3.
Cytokine/Chemokine Release of PHA/PMA-Stimulated Jurkat Human T-Cell Leukemia Cells and TNFα-Stimulated A549 Cells
| ExcelArray Inflammation I | IFNγ | IL-1β | IL-2 | IL-4 | IL-6 | IL-8 | IL-10 | IL-12p70 | RANTES | MIP-1α | MIP-1β | TNFα |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Jurkat + PHA/PMA | — | — | >1000 | — | — | >1000 | — | — | — | >1000 | 169 | 770 |
| A549 + TNFα | — | — | — | — | 80 | 26,000 | — | — | 17,000 | — | 50 | — |
| ExcelArray Inflammation II | IL-1α | IL-3 | IL-5 | IL-7 | IL-13 | IL-15 | IL-17 | FasL | G-CSF | GM- CSF | TNFβ | MCP-1 |
| Jurkat + PHA/PMA | — | >1000 | — | — | 400 | — | — | 40 | 105 | >1000 | 419 | — |
| A549 + TNFα | — | — | — | — | — | — | — | — | 120 | — | — | 18,000 |
| ExcelArray Angiogenesis | Angiogenin | EGF | FGFb | GROα | HGF | IL-8 | HB- EGF | KGF | PlGF | VEGF | TIMP-1 | TNFα |
| A549 + TNFα | — | — | — | 38,000 | — | 26,000 | — | — | — | — | 21,000 | — |
| ExcelArray Chemotaxis | Eotaxin | I-309 | IP-10 | MCP-3 | MDC | IL-8 | Fractalkine | TARC | RANTES | MIP-1α | MIP-1β | MCP-1 |
| Jurkat + PHA/PMA | — | >1000 | 93 | — | 95 | >1000 | — | — | — | >1000 | 169 | — |
| A549 + TNFα | — | — | 490 | — | 250 | 26,000 | — | 130 | 17,000 | — | 50 | 18,000 |
Analytes detected at an increased level in stimulated cell culture supernatants as compared to non-stimulated controls are listed. Values listed are the difference of stimulated cell culture supernatant analyte concentration minus non-stimulated cell culture supernatant analyte concentration.
FIGURE 3.
Representative microarray images of (A) nonstimulated A549 cell-culture supernatants and (B) TNFα-stimulated A549 cell-culture supernatants.
In conclusion, the use of Thermo Scientific Excel-Array antibody microarrays in these experiments enabled the simultaneous screening of 41 human cytokines and chemokines in experimental samples. These microarrays were used to rapidly analyze cytokine/chemokine levels in stimulated and nonstimulated Jurkat and A549 human cell-culture supernatants. The multiplexing capability of ExcelArray human microarrays results in time and sample savings over traditional ELISA methods. Furthermore, the ExcelArray microarrays contain thoroughly screened antibody pairs, proven slide printing and post-printing treatment parameters, and optimized protocols that are capable of detecting naturally expressed targets in model biological systems. These attributes yield a superior tool for cytokine/chemokine analysis as compared to traditional ELISA.
REFERENCES
- 1.Collins FS. Microarrays and macroconsequences. Nature Genet. 1999;21:2. [Google Scholar]
- 2.Lander ES. Array of hope. Nature Genet. 1999;21:3–4. doi: 10.1038/4427. [DOI] [PubMed] [Google Scholar]
- 3.Southern E, Mir K, Shchepinov M. Molecular interactions on arrays. Nature Genet. 1999;21:5–9. doi: 10.1038/4429. [DOI] [PubMed] [Google Scholar]
- 4.Mendoza LG, McQuarry P, Mongan A, Gangadharan R, Brignac S, Eggers M. High-throughput microarray based enzyme-linked immunosorbent assay. Biotechniques. 1999;27:778–788. doi: 10.2144/99274rr01. [DOI] [PubMed] [Google Scholar]
- 5.Ekins RP, Chu FW. Microarrays: Their origins and applications. Trends Biotechnol. 1999;17:217–218. doi: 10.1016/s0167-7799(99)01329-3. [DOI] [PubMed] [Google Scholar]
- 6.Lueking C, Horn M, Eickhoff H, Bussow K, Lehrach H, Walker G. Protein microarrays for gene expression and antibody screening. Anal Biochem. 1999;270:103–111. doi: 10.1006/abio.1999.4063. [DOI] [PubMed] [Google Scholar]
- 7.Haab BB, Dunham MJ, Brown PO. Protein microarrays for highly parallel detection and quantitation of specific proteins and antibodies in complex solutions. Genome Biol. 2001;2(2):Research 0004.1–0004.13. doi: 10.1186/gb-2001-2-2-research0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiese R, Belosludtsev Y, Powdrill T, Thompson P, Hogan M. Simultaneous multianalyte ELISA performed on a microarray platform. Clin Chem. 2001;47:1451–1457. [PubMed] [Google Scholar]
- 9.Tam S, Wiese R, Lee S, Gilmore J, Kumble KD. Simultaneous analysis of eight human Th1/Th2 cytokines using microarrays. J. Immunol Methods. 2002;261:157–165. doi: 10.1016/s0022-1759(01)00572-5. [DOI] [PubMed] [Google Scholar]
- 10.Gerlag D, Ransone L, Tak P, Han Z, Palanki M, Barbosa M, et al. The effect of a T-cell-specific NF-κB inhibitor on in vitro cytokine production and collagen-induced arthritis. J Immunol. 2000;165:1652–1658. doi: 10.4049/jimmunol.165.3.1652. [DOI] [PubMed] [Google Scholar]
- 11.Osawa Y, Nagaki M, Banno Y, Brenner D, Asano T, Nozawa Y, et al. Tumor necrosis factor alpha–induced interleukin-8 production via NF-κB and phosphatidylinositol 3-kinase/Akt pathways inhibits cell apoptosis in human heptocytes. Infection and Immunity. 2002;70:6294–6301. doi: 10.1128/IAI.70.11.6294-6301.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erdelyi K, Kiss A, Bakondi E, Bai P, Szabo C, Gergely P, et al. Gallotanin inhibits the expression of chemokines and inflammatory cytokines in A549 cells. Mol Pharm. 2005;68:895–904. doi: 10.1124/mol.105.012518. [DOI] [PubMed] [Google Scholar]