Abstract
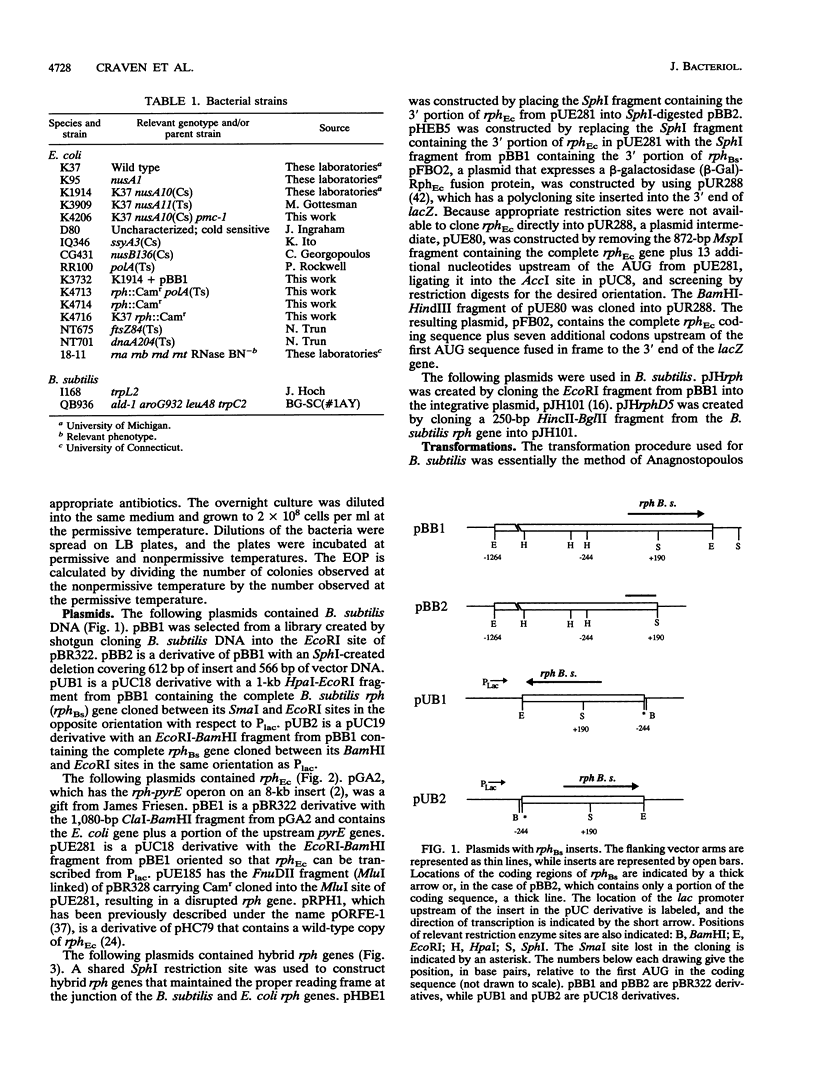
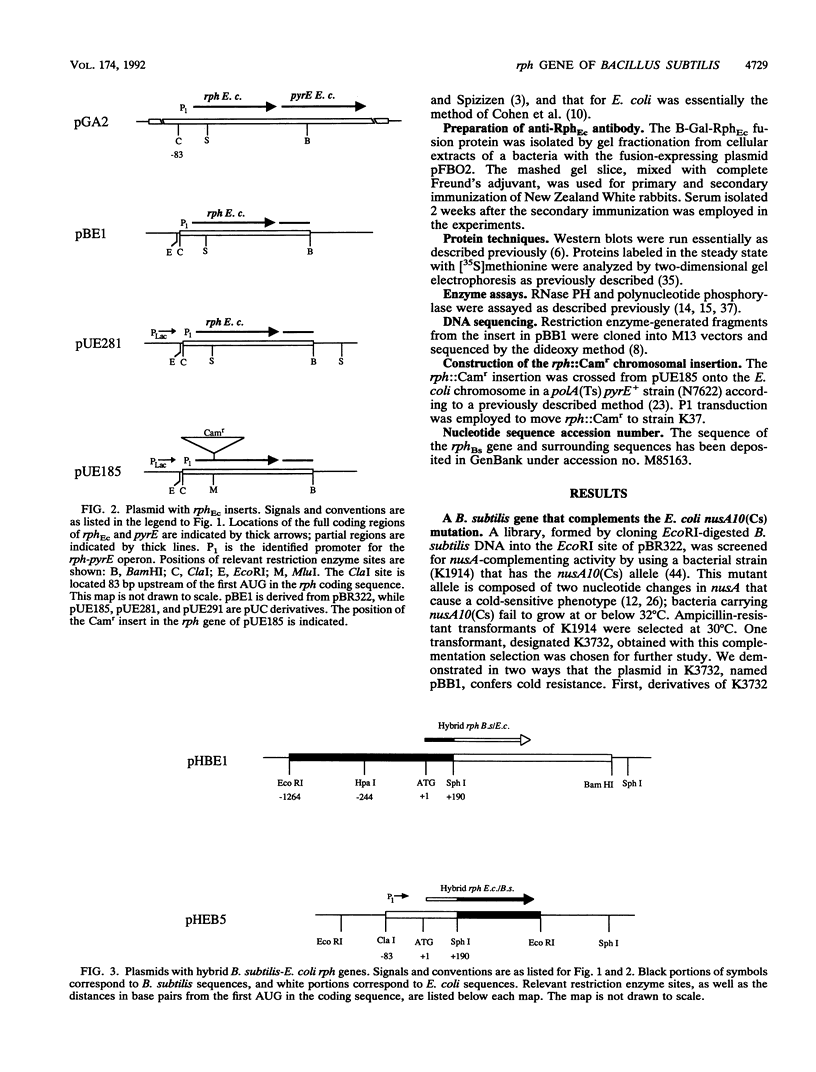
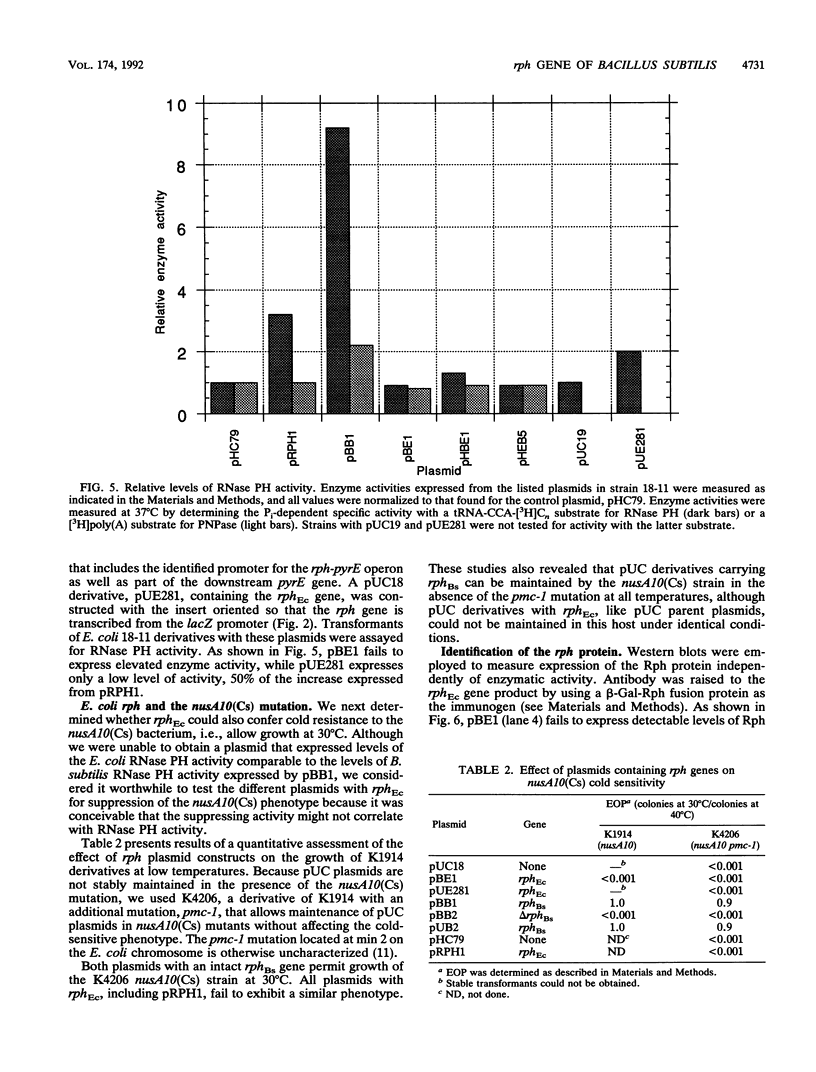
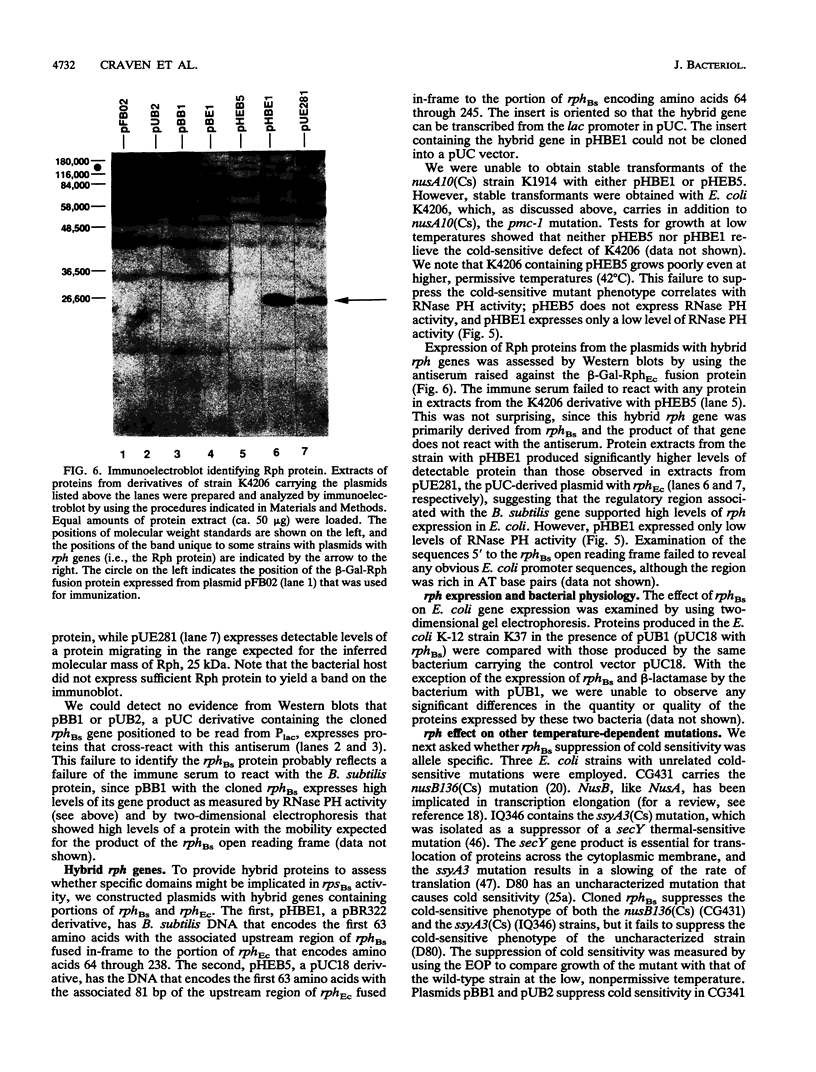
A shotgun cloning of Bacillus subtilis DNA into pBR322 yielded a 2-kb fragment that suppresses the cold-sensitive defect of the nusA10(Cs) Escherichia coli mutant. The responsible gene encodes an open reading frame that is greater than 50% identical at the amino acid level to the E. coli rph gene, which was formerly called orfE. This B. subtilis gene is located at 251 degrees adjacent to the gerM gene on the B. subtilis genetic map. It has been named rph because, like its E. coli analog, it encodes a phosphate-dependent exoribonuclease activity, RNase PH, that removes the 3' nucleotides from precursor tRNAs. The cloned B. subtilis rph gene also suppresses the cold-sensitive phenotype of other unrelated cold-sensitive mutants of E. coli, but not the temperature-sensitive phenotype of three temperature-sensitive mutants, including the nusA11(Ts) mutant, that were tested.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albertini A. M., Galizzi A. The Bacillus subtilis outB gene is highly homologous to an Escherichia coli ntr-like gene. J Bacteriol. 1990 Sep;172(9):5482–5485. doi: 10.1128/jb.172.9.5482-5485.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An G., Justesen J., Watson R. J., Friesen J. D. Cloning the spoT gene of Escherichia coli: identification of the spoT gene product. J Bacteriol. 1979 Mar;137(3):1100–1110. doi: 10.1128/jb.137.3.1100-1110.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anagnostopoulos C., Spizizen J. REQUIREMENTS FOR TRANSFORMATION IN BACILLUS SUBTILIS. J Bacteriol. 1961 May;81(5):741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen J. T., Jensen K. F., Poulsen P. Role of transcription pausing in the control of the pyrE attenuator in Escherichia coli. Mol Microbiol. 1991 Feb;5(2):327–333. doi: 10.1111/j.1365-2958.1991.tb02113.x. [DOI] [PubMed] [Google Scholar]
- Aranki A., Freter R. Use of anaerobic glove boxes for the cultivation of strictly anaerobic bacteria. Am J Clin Nutr. 1972 Dec;25(12):1329–1334. doi: 10.1093/ajcn/25.12.1329. [DOI] [PubMed] [Google Scholar]
- Beall B., Lowe M., Lutkenhaus J. Cloning and characterization of Bacillus subtilis homologs of Escherichia coli cell division genes ftsZ and ftsA. J Bacteriol. 1988 Oct;170(10):4855–4864. doi: 10.1128/jb.170.10.4855-4864.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonekamp F., Andersen H. D., Christensen T., Jensen K. F. Codon-defined ribosomal pausing in Escherichia coli detected by using the pyrE attenuator to probe the coupling between transcription and translation. Nucleic Acids Res. 1985 Jun 11;13(11):4113–4123. doi: 10.1093/nar/13.11.4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven M. G., Friedman D. I. Analysis of the Escherichia coli nusA10(Cs) allele: relating nucleotide changes to phenotypes. J Bacteriol. 1991 Feb;173(4):1485–1491. doi: 10.1128/jb.173.4.1485-1491.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cudny H., Deutscher M. P. 3' processing of tRNA precursors in ribonuclease-deficient Escherichia coli. Development and characterization of an in vitro processing system and evidence for a phosphate requirement. J Biol Chem. 1988 Jan 25;263(3):1518–1523. [PubMed] [Google Scholar]
- Deutscher M. P., Ghosh R. K. Preparation of synthetic tRNA precursors with tRNA nucleotidyltransferase. Nucleic Acids Res. 1978 Oct;5(10):3821–3829. doi: 10.1093/nar/5.10.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher M. P., Marshall G. T., Cudny H. RNase PH: an Escherichia coli phosphate-dependent nuclease distinct from polynucleotide phosphorylase. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4710–4714. doi: 10.1073/pnas.85.13.4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari F. A., Nguyen A., Lang D., Hoch J. A. Construction and properties of an integrable plasmid for Bacillus subtilis. J Bacteriol. 1983 Jun;154(3):1513–1515. doi: 10.1128/jb.154.3.1513-1515.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman D. I. Integration host factor: a protein for all reasons. Cell. 1988 Nov 18;55(4):545–554. doi: 10.1016/0092-8674(88)90213-9. [DOI] [PubMed] [Google Scholar]
- Friedman D. I., Jolly C. T., Mural R. J. Interference with the expression of the N gene function of phage lambda in a mutant of Escherichia coli. Virology. 1973 Jan;51(1):216–226. doi: 10.1016/0042-6822(73)90381-4. [DOI] [PubMed] [Google Scholar]
- Georgopoulos C. P., Swindle J., Keppel F., Ballivet M., Bisig R., Eisen H. Studies on the E. coli groNB (nusB) gene which affects bacteriophage lambda N gene function. Mol Gen Genet. 1980;179(1):55–61. doi: 10.1007/BF00268446. [DOI] [PubMed] [Google Scholar]
- Grundy F. J., Henkin T. M. Cloning and analysis of the Bacillus subtilis rpsD gene, encoding ribosomal protein S4. J Bacteriol. 1990 Nov;172(11):6372–6379. doi: 10.1128/jb.172.11.6372-6379.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C., Nashimoto H., Nomura M. Structure and function of E. coli ribosomes. 8. Cold-sensitive mutants defective in ribosome assembly. Proc Natl Acad Sci U S A. 1969 Jun;63(2):384–391. doi: 10.1073/pnas.63.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutterson N. I., Koshland D. E., Jr Replacement and amplification of bacterial genes with sequences altered in vitro. Proc Natl Acad Sci U S A. 1983 Aug;80(16):4894–4898. doi: 10.1073/pnas.80.16.4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch J. A., Barat M., Anagnostopoulos C. Transformation and transduction in recombination-defective mutants of Bacillus subtilis. J Bacteriol. 1967 Jun;93(6):1925–1937. doi: 10.1128/jb.93.6.1925-1937.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohn B., Collins J. A small cosmid for efficient cloning of large DNA fragments. Gene. 1980 Nov;11(3-4):291–298. doi: 10.1016/0378-1119(80)90069-4. [DOI] [PubMed] [Google Scholar]
- Ito K., Egawa K., Nakamura Y. Genetic interaction between the beta' subunit of RNA polymerase and the arginine-rich domain of Escherichia coli nusA protein. J Bacteriol. 1991 Feb;173(4):1492–1501. doi: 10.1128/jb.173.4.1492-1501.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen K. F., Pedersen S. Metabolic growth rate control in Escherichia coli may be a consequence of subsaturation of the macromolecular biosynthetic apparatus with substrates and catalytic components. Microbiol Rev. 1990 Jun;54(2):89–100. doi: 10.1128/mr.54.2.89-100.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- Moir D., Stewart S. E., Osmond B. C., Botstein D. Cold-sensitive cell-division-cycle mutants of yeast: isolation, properties, and pseudoreversion studies. Genetics. 1982 Apr;100(4):547–563. doi: 10.1093/genetics/100.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y., Mizusawa S., Court D. L., Tsugawa A. Regulatory defects of a conditionally lethal nusAts mutant of Escherichia coli. Positive and negative modulator roles of NusA protein in vivo. J Mol Biol. 1986 May 5;189(1):103–111. doi: 10.1016/0022-2836(86)90384-0. [DOI] [PubMed] [Google Scholar]
- Neuhard J., Stauning E., Kelln R. A. Cloning and characterization of the pyrE gene and of PyrE::Mud1 (Ap lac) fusions from Salmonella typhimurium. Eur J Biochem. 1985 Feb 1;146(3):597–603. doi: 10.1111/j.1432-1033.1985.tb08693.x. [DOI] [PubMed] [Google Scholar]
- Novick P., Osmond B. C., Botstein D. Suppressors of yeast actin mutations. Genetics. 1989 Apr;121(4):659–674. doi: 10.1093/genetics/121.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Farrell P. H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975 May 25;250(10):4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Ost K. A., Deutscher M. P. Escherichia coli orfE (upstream of pyrE) encodes RNase PH. J Bacteriol. 1991 Sep;173(17):5589–5591. doi: 10.1128/jb.173.17.5589-5591.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ost K. A., Deutscher M. P. RNase PH catalyzes a synthetic reaction, the addition of nucleotides to the 3' end of RNA. Biochimie. 1990 Nov;72(11):813–818. doi: 10.1016/0300-9084(90)90190-r. [DOI] [PubMed] [Google Scholar]
- Parker J. Errors and alternatives in reading the universal genetic code. Microbiol Rev. 1989 Sep;53(3):273–298. doi: 10.1128/mr.53.3.273-298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen P., Andersen J. T., Jensen K. F. Molecular and mutational analysis of three genes preceding pyrE on the Escherichia coli chromosome. Mol Microbiol. 1989 Mar;3(3):393–404. doi: 10.1111/j.1365-2958.1989.tb00184.x. [DOI] [PubMed] [Google Scholar]
- Poulsen P., Bonekamp F., Jensen K. F. Structure of the Escherichia coli pyrE operon and control of pyrE expression by a UTP modulated intercistronic attentuation. EMBO J. 1984 Aug;3(8):1783–1790. doi: 10.1002/j.1460-2075.1984.tb02046.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J. W. Phage lambda and the regulation of transcription termination. Cell. 1988 Jan 15;52(1):5–6. doi: 10.1016/0092-8674(88)90523-5. [DOI] [PubMed] [Google Scholar]
- Rüther U., Müller-Hill B. Easy identification of cDNA clones. EMBO J. 1983;2(10):1791–1794. doi: 10.1002/j.1460-2075.1983.tb01659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sammons R. L., Slynn G. M., Smith D. A. Genetical and molecular studies on gerM, a new developmental locus of Bacillus subtilis. J Gen Microbiol. 1987 Dec;133(12):3299–3312. doi: 10.1099/00221287-133-12-3299. [DOI] [PubMed] [Google Scholar]
- Schauer A. T., Carver D. L., Bigelow B., Baron L. S., Friedman D. I. lambda N antitermination system: functional analysis of phage interactions with the host NusA protein. J Mol Biol. 1987 Apr 20;194(4):679–690. doi: 10.1016/0022-2836(87)90245-2. [DOI] [PubMed] [Google Scholar]
- Shazand K., Tucker J., Chiang R., Stansmore K., Sperling-Petersen H. U., Grunberg-Manago M., Rabinowitz J. C., Leighton T. Isolation and molecular genetic characterization of the Bacillus subtilis gene (infB) encoding protein synthesis initiation factor 2. J Bacteriol. 1990 May;172(5):2675–2687. doi: 10.1128/jb.172.5.2675-2687.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba K., Ito K., Yura T., Cerretti D. P. A defined mutation in the protein export gene within the spc ribosomal protein operon of Escherichia coli: isolation and characterization of a new temperature-sensitive secY mutant. EMBO J. 1984 Mar;3(3):631–635. doi: 10.1002/j.1460-2075.1984.tb01859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba K., Ito K., Yura T. Mutation that suppresses the protein export defect of the secY mutation and causes cold-sensitive growth of Escherichia coli. J Bacteriol. 1984 Nov;160(2):696–701. doi: 10.1128/jb.160.2.696-701.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai P. C., Kessler D. P., Ingraham J. Cold-sensitive mutations in Salmonella typhimurium which affect ribosome synthesis. J Bacteriol. 1969 Mar;97(3):1298–1304. doi: 10.1128/jb.97.3.1298-1304.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]




