Abstract
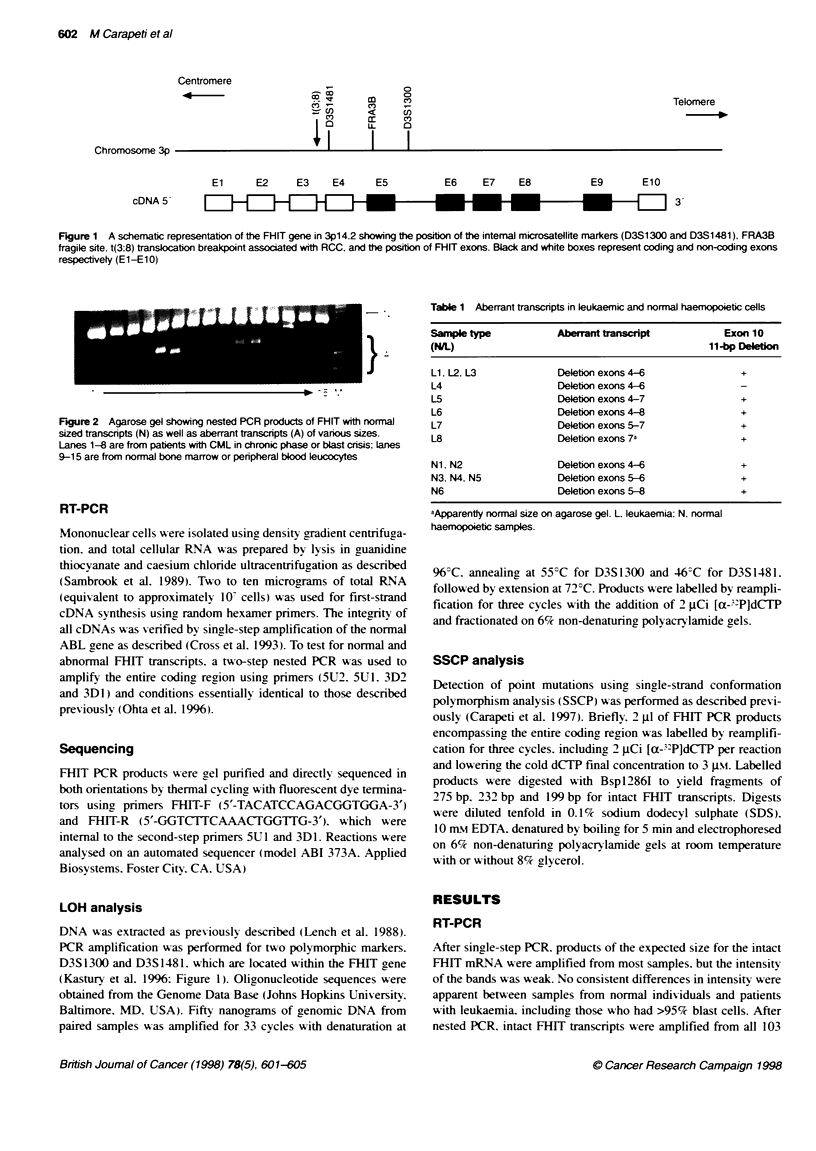
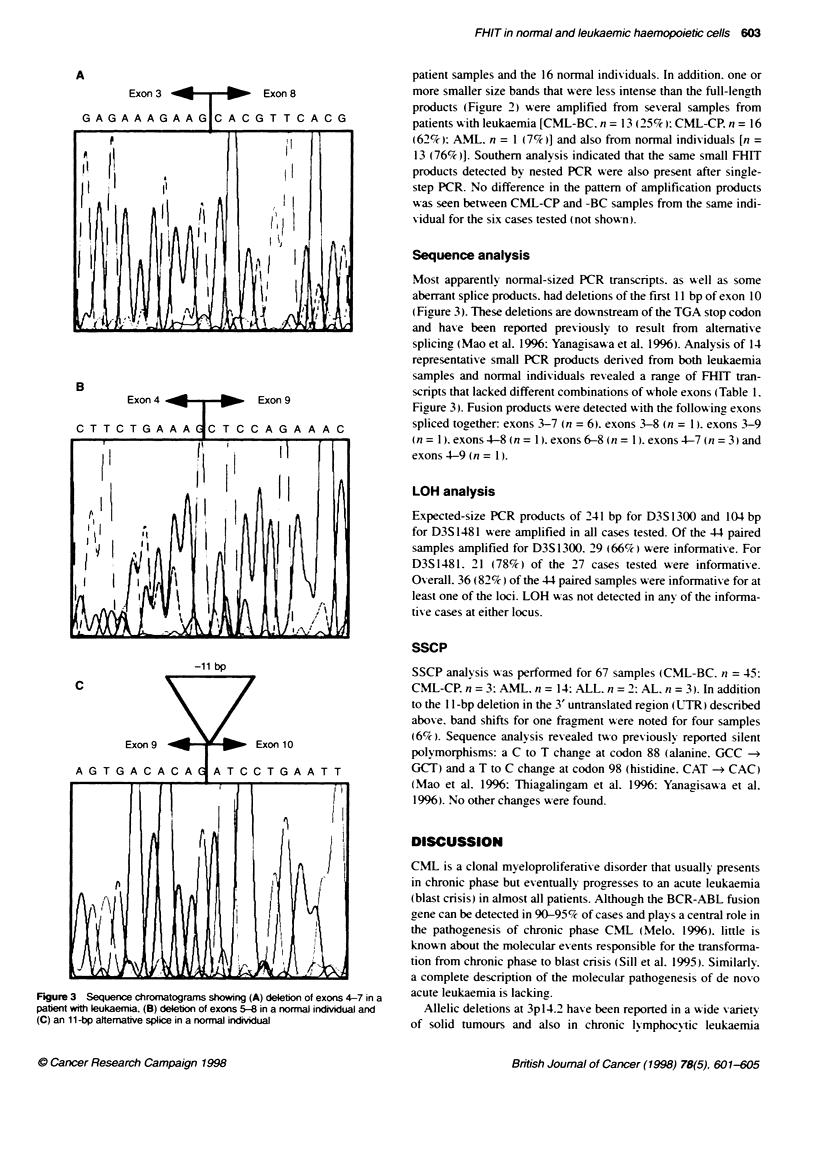
Deletions and apparent transcriptional abnormalities of the FHIT gene at 3p14.2 have recently been reported in a wide variety of solid tumours. To determine whether lesions of this gene also occur in leukaemia, we have analysed a total of 97 patients (chronic myeloid leukaemia, CML, in chronic phase or blast crisis, n = 71; de novo acute leukaemia, n = 26) and 16 normal individuals. Intact FHIT transcripts from all cases were amplified using RT-PCR. In addition, smaller size bands that were less intense than the full-length products were amplified from several samples from patients with leukaemia and also from normal leucocytes. Sequencing of the small products revealed that they were derived from FHIT transcripts lacking whole exons. Using single-strand conformation polymorphism analysis, no mutations in the coding sequence were detected in any patient. Furthermore, loss of heterozygosity was not seen in any of 36 informative patients at D3S1300 or D3S1481, markers located within the FHIT locus. We conclude that the FHIT gene and other uncharacterized tumour-suppressor genes at 3p14.2 are unlikely to be involved in the pathogenesis of acute leukaemia or progression of CML from chronic phase to blast crisis. Moreover, low-abundance FHIT transcripts that lack whole exons are not specific to malignant cells and should not be taken as evidence of an abnormality in the absence of demonstrable genomic DNA lesions.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barnes L. D., Garrison P. N., Siprashvili Z., Guranowski A., Robinson A. K., Ingram S. W., Croce C. M., Ohta M., Huebner K. Fhit, a putative tumor suppressor in humans, is a dinucleoside 5',5"'-P1,P3-triphosphate hydrolase. Biochemistry. 1996 Sep 10;35(36):11529–11535. doi: 10.1021/bi961415t. [DOI] [PubMed] [Google Scholar]
- Brauch H., Johnson B., Hovis J., Yano T., Gazdar A., Pettengill O. S., Graziano S., Sorenson G. D., Poiesz B. J., Minna J. Molecular analysis of the short arm of chromosome 3 in small-cell and non-small-cell carcinoma of the lung. N Engl J Med. 1987 Oct 29;317(18):1109–1113. doi: 10.1056/NEJM198710293171803. [DOI] [PubMed] [Google Scholar]
- Carapeti M., Goldman J. M., Cross N. C. Dominant-negative mutations of the Wilms' tumour predisposing gene (WT1) are infrequent in CML blast crisis and de novo acute leukaemia. Eur J Haematol. 1997 May;58(5):346–349. doi: 10.1111/j.1600-0609.1997.tb01681.x. [DOI] [PubMed] [Google Scholar]
- Cohen A. J., Li F. P., Berg S., Marchetto D. J., Tsai S., Jacobs S. C., Brown R. S. Hereditary renal-cell carcinoma associated with a chromosomal translocation. N Engl J Med. 1979 Sep 13;301(11):592–595. doi: 10.1056/NEJM197909133011107. [DOI] [PubMed] [Google Scholar]
- Gemma A., Hagiwara K., Ke Y., Burke L. M., Khan M. A., Nagashima M., Bennett W. P., Harris C. C. FHIT mutations in human primary gastric cancer. Cancer Res. 1997 Apr 15;57(8):1435–1437. [PubMed] [Google Scholar]
- Hayashi S., Tanimoto K., Hajiro-Nakanishi K., Tsuchiya E., Kurosumi M., Higashi Y., Imai K., Suga K., Nakachi K. Abnormal FHIT transcripts in human breast carcinomas: a clinicopathological and epidemiological analysis of 61 Japanese cases. Cancer Res. 1997 May 15;57(10):1981–1985. [PubMed] [Google Scholar]
- Hendricks D. T., Taylor R., Reed M., Birrer M. J. FHIT gene expression in human ovarian, endometrial, and cervical cancer cell lines. Cancer Res. 1997 Jun 1;57(11):2112–2115. [PubMed] [Google Scholar]
- Kastury K., Baffa R., Druck T., Ohta M., Cotticelli M. G., Inoue H., Negrini M., Rugge M., Huang D., Croce C. M. Potential gastrointestinal tumor suppressor locus at the 3p14.2 FRA3B site identified by homozygous deletions in tumor cell lines. Cancer Res. 1996 Mar 1;56(5):978–983. [PubMed] [Google Scholar]
- Kok K., Naylor S. L., Buys C. H. Deletions of the short arm of chromosome 3 in solid tumors and the search for suppressor genes. Adv Cancer Res. 1997;71:27–92. doi: 10.1016/s0065-230x(08)60096-2. [DOI] [PubMed] [Google Scholar]
- Lench N., Stanier P., Williamson R. Simple non-invasive method to obtain DNA for gene analysis. Lancet. 1988 Jun 18;1(8599):1356–1358. doi: 10.1016/s0140-6736(88)92178-2. [DOI] [PubMed] [Google Scholar]
- Lisitsyn N. A., Leach F. S., Vogelstein B., Wigler M. H. Detection of genetic loss in tumors by representational difference analysis. Cold Spring Harb Symp Quant Biol. 1994;59:585–587. doi: 10.1101/sqb.1994.059.01.066. [DOI] [PubMed] [Google Scholar]
- Luan X., Shi G., Zohouri M., Paradee W., Smith D. I., Decker H. J., Cannizzaro L. A. The FHIT gene is alternatively spliced in normal kidney and renal cell carcinoma. Oncogene. 1997 Jul 3;15(1):79–86. doi: 10.1038/sj.onc.1201164. [DOI] [PubMed] [Google Scholar]
- Man S., Ellis I. O., Sibbering M., Blamey R. W., Brook J. D. High levels of allele loss at the FHIT and ATM genes in non-comedo ductal carcinoma in situ and grade I tubular invasive breast cancers. Cancer Res. 1996 Dec 1;56(23):5484–5489. [PubMed] [Google Scholar]
- Mao L., Fan Y. H., Lotan R., Hong W. K. Frequent abnormalities of FHIT, a candidate tumor suppressor gene, in head and neck cancer cell lines. Cancer Res. 1996 Nov 15;56(22):5128–5131. [PubMed] [Google Scholar]
- Melo J. V. The diversity of BCR-ABL fusion proteins and their relationship to leukemia phenotype. Blood. 1996 Oct 1;88(7):2375–2384. [PubMed] [Google Scholar]
- Naylor S. L., Johnson B. E., Minna J. D., Sakaguchi A. Y. Loss of heterozygosity of chromosome 3p markers in small-cell lung cancer. Nature. 1987 Oct 1;329(6138):451–454. doi: 10.1038/329451a0. [DOI] [PubMed] [Google Scholar]
- Negrini M., Monaco C., Vorechovsky I., Ohta M., Druck T., Baffa R., Huebner K., Croce C. M. The FHIT gene at 3p14.2 is abnormal in breast carcinomas. Cancer Res. 1996 Jul 15;56(14):3173–3179. [PubMed] [Google Scholar]
- Ohta M., Inoue H., Cotticelli M. G., Kastury K., Baffa R., Palazzo J., Siprashvili Z., Mori M., McCue P., Druck T. The FHIT gene, spanning the chromosome 3p14.2 fragile site and renal carcinoma-associated t(3;8) breakpoint, is abnormal in digestive tract cancers. Cell. 1996 Feb 23;84(4):587–597. doi: 10.1016/s0092-8674(00)81034-x. [DOI] [PubMed] [Google Scholar]
- Panagopoulos I., Pandis N., Thelin S., Petersson C., Mertens F., Borg A., Kristoffersson U., Mitelman F., Aman P. The FHIT and PTPRG genes are deleted in benign proliferative breast disease associated with familial breast cancer and cytogenetic rearrangements of chromosome band 3p14. Cancer Res. 1996 Nov 1;56(21):4871–4875. [PubMed] [Google Scholar]
- Panagopoulos I., Thelin S., Mertens F., Mitelman F., Aman P. Variable FHIT transcripts in non-neoplastic tissues. Genes Chromosomes Cancer. 1997 Aug;19(4):215–219. doi: 10.1002/(sici)1098-2264(199708)19:4<215::aid-gcc2>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Paradee W., Mullins C., He Z., Glover T., Wilke C., Opalka B., Schutte J., Smith D. I. Precise localization of aphidicolin-induced breakpoints on the short arm of human chromosome 3. Genomics. 1995 May 20;27(2):358–361. doi: 10.1006/geno.1995.1057. [DOI] [PubMed] [Google Scholar]
- Shridhar R., Shridhar V., Wang X., Paradee W., Dugan M., Sarkar F., Wilke C., Glover T. W., Vaitkevicius V. K., Smith D. I. Frequent breakpoints in the 3p14.2 fragile site, FRA3B, in pancreatic tumors. Cancer Res. 1996 Oct 1;56(19):4347–4350. [PubMed] [Google Scholar]
- Sill H., Goldman J. M., Cross N. C. Homozygous deletions of the p16 tumor-suppressor gene are associated with lymphoid transformation of chronic myeloid leukemia. Blood. 1995 Apr 15;85(8):2013–2016. [PubMed] [Google Scholar]
- Sozzi G., Alder H., Tornielli S., Corletto V., Baffa R., Veronese M. L., Negrini M., Pilotti S., Pierotti M. A., Huebner K. Aberrant FHIT transcripts in Merkel cell carcinoma. Cancer Res. 1996 Jun 1;56(11):2472–2474. [PubMed] [Google Scholar]
- Sozzi G., Veronese M. L., Negrini M., Baffa R., Cotticelli M. G., Inoue H., Tornielli S., Pilotti S., De Gregorio L., Pastorino U. The FHIT gene 3p14.2 is abnormal in lung cancer. Cell. 1996 Apr 5;85(1):17–26. doi: 10.1016/s0092-8674(00)81078-8. [DOI] [PubMed] [Google Scholar]
- Thiagalingam S., Lisitsyn N. A., Hamaguchi M., Wigler M. H., Willson J. K., Markowitz S. D., Leach F. S., Kinzler K. W., Vogelstein B. Evaluation of the FHIT gene in colorectal cancers. Cancer Res. 1996 Jul 1;56(13):2936–2939. [PubMed] [Google Scholar]
- Virgilio L., Shuster M., Gollin S. M., Veronese M. L., Ohta M., Huebner K., Croce C. M. FHIT gene alterations in head and neck squamous cell carcinomas. Proc Natl Acad Sci U S A. 1996 Sep 3;93(18):9770–9775. doi: 10.1073/pnas.93.18.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa K., Kondo M., Osada H., Uchida K., Takagi K., Masuda A., Takahashi T., Takahashi T. Molecular analysis of the FHIT gene at 3p14.2 in lung cancer cell lines. Cancer Res. 1996 Dec 15;56(24):5579–5582. [PubMed] [Google Scholar]
- Zbar B., Brauch H., Talmadge C., Linehan M. Loss of alleles of loci on the short arm of chromosome 3 in renal cell carcinoma. 1987 Jun 25-Jul 1Nature. 327(6124):721–724. doi: 10.1038/327721a0. [DOI] [PubMed] [Google Scholar]
- Zeiger M. A., Gnarra J. R., Zbar B., Linehan W. M., Pass H. I. Loss of heterozygosity on the short arm of chromosome 3 in mesothelioma cell lines and solid tumors. Genes Chromosomes Cancer. 1994 Sep;11(1):15–20. doi: 10.1002/gcc.2870110104. [DOI] [PubMed] [Google Scholar]
- Zeiger M. A., Zbar B., Keiser H., Linehan W. M., Gnarra J. R. Loss of heterozygosity on the short arm of chromosome 3 in sporadic, von Hippel-Lindau disease-associated, and familial pheochromocytoma. Genes Chromosomes Cancer. 1995 Jul;13(3):151–156. doi: 10.1002/gcc.2870130303. [DOI] [PubMed] [Google Scholar]
- Zou T. T., Lei J., Shi Y. Q., Yin J., Wang S., Souza R. F., Kong D., Shimada Y., Smolinski K. N., Greenwald B. D. FHIT gene alterations in esophageal cancer and ulcerative colitis (UC). Oncogene. 1997 Jul 3;15(1):101–105. doi: 10.1038/sj.onc.1201169. [DOI] [PubMed] [Google Scholar]
- van den Berg A., Draaijers T. G., Kok K., Timmer T., Van der Veen A. Y., Veldhuis P. M., de Leij L., Gerhartz C. D., Naylor S. L., Smith D. I. Normal FHIT transcripts in renal cell cancer- and lung cancer-derived cell lines, including a cell line with a homozygous deletion in the FRA3B region. Genes Chromosomes Cancer. 1997 Aug;19(4):220–227. doi: 10.1002/(sici)1098-2264(199708)19:4<220::aid-gcc3>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]



