Abstract
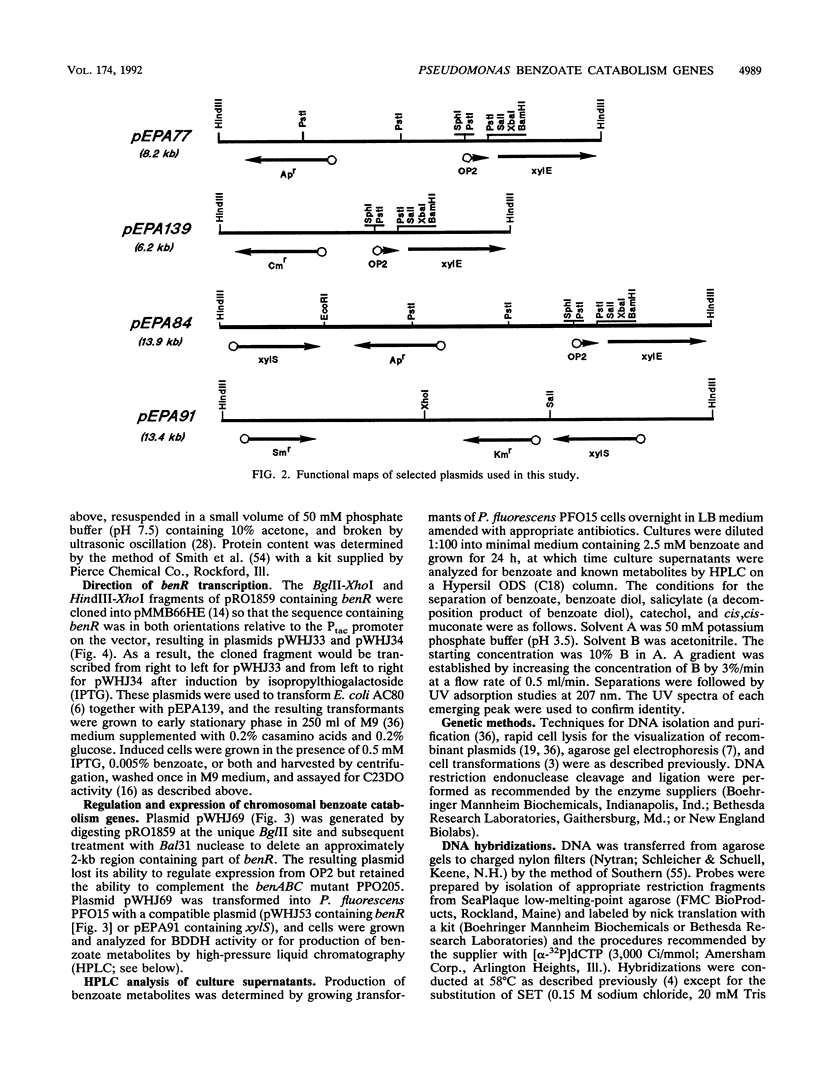
Mutants of Pseudomonas putida mt-2 that are unable to convert benzoate to catechol were isolated and grouped into two classes: those that did not initiate attack on benzoate and those that accumulated 3,5-cyclohexadiene-1,2-diol-1-carboxylic acid (benzoate diol). The latter mutants, represents by strain PP0201, were shown to lack benzoate diol dehydrogenase (benD) activity. Mutants from the former class were presumed either to carry lesions in one or more subunit structural genes of benzoate dioxygenase (benABC) or the regulatory gene (benR) or to contain multiple mutations. Previous work in this laboratory suggested that benR can substitute for the TOL plasmid-encoded xylS regulatory gene, which promotes gene expression from the OP2 region of the lower or meta pathway operon. Accordingly, structural and regulatory gene mutations were distinguished by the ability of benzoate-grown mutant strains to induce expression from OP2 without xylS by using the TOL plasmid xylE gene (encoding catechol 2,3-dioxygenase) as a reporter. A cloned 12-kb BamHI chromosomal DNA fragment from the P. aeruginosa PAO1 chromosome complemented all of the mutations, as shown by restoration of growth on benzoate minimal medium. Subcloning and deletion analyses allowed identification of DNA fragments carrying benD, benABC, and the region possessing xylS substitution activity, benR. Expression of these genes was examined in a strain devoid of benzoate-utilizing ability, Pseudomonas fluorescens PFO15. The disappearance of benzoate and the production of catechol were determined by chromatographic analysis of supernatants from cultures grown with casamino acids. When P. fluorescens PFO15 was transformed with plasmids containing only benABCD, no loss of benzoate was observed. When either benR or xylS was cloned into plasmids compatible with those plasmids containing only the benABCD regions, benzoate was removed from the medium and catechol was produced. Regulation of expression of the chromosomal structural genes by benR and xylS was quantified by benzoate diol dehydrogenase enzyme assays. The results obtained when xylS was substituted for benR strongly suggest an isofunctional regulatory mechanism between the TOL plasmid lower-pathway genes (via the OP2 promoter) and chromosomal benABC. Southern hybridizations demonstrated that DNA encoding the benzoate dioxygenase structural genes showed homology to DNA encoding toluate dioxygenase from the TOL plasmid pWW0, but benR did not show homology to xylS. Evolutionary relationships between the regulatory systems of chromosomal and plasmid-encoded genes for the catabolism of benzoate and related compounds are suggested.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aldrich T. L., Frantz B., Gill J. F., Kilbane J. J., Chakrabarty A. M. Cloning and complete nucleotide sequence determination of the catB gene encoding cis,cis-muconate lactonizing enzyme. Gene. 1987;52(2-3):185–195. doi: 10.1016/0378-1119(87)90045-x. [DOI] [PubMed] [Google Scholar]
- Axcell B. C., Geary P. J. Purification and some properties of a soluble benzene-oxidizing system from a strain of Pseudomonas. Biochem J. 1975 Jan;146(1):173–183. doi: 10.1042/bj1460173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagdasarian M., Timmis K. N. Host: vector systems for gene cloning in Pseudomonas. Curr Top Microbiol Immunol. 1982;96:47–67. doi: 10.1007/978-3-642-68315-2_4. [DOI] [PubMed] [Google Scholar]
- Barkay T., Fouts D. L., Olson B. H. Preparation of a DNA gene probe for detection of mercury resistance genes in gram-negative bacterial communities. Appl Environ Microbiol. 1985 Mar;49(3):686–692. doi: 10.1128/aem.49.3.686-692.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuskey S. M., Peccoraro V., Olsen R. H. Initial catabolism of aromatic biogenic amines by Pseudomonas aeruginosa PAO: pathway description, mapping of mutations, and cloning of essential genes. J Bacteriol. 1987 Jun;169(6):2398–2404. doi: 10.1128/jb.169.6.2398-2404.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuskey S. M., Sprenkle A. B. Benzoate-dependent induction from the OP2 operator-promoter region of the TOL plasmid pWWO in the absence of known plasmid regulatory genes. J Bacteriol. 1988 Aug;170(8):3742–3746. doi: 10.1128/jb.170.8.3742-3746.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuskey S. M., Wolff J. A., Phibbs P. V., Jr, Olsen R. H. Cloning of genes specifying carbohydrate catabolism in Pseudomonas aeruginosa and Pseudomonas putida. J Bacteriol. 1985 Jun;162(3):865–871. doi: 10.1128/jb.162.3.865-871.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison J., Heusterspreute M., Chevalier N., Ha-Thi V., Brunel F. Vectors with restriction site banks. V. pJRD215, a wide-host-range cosmid vector with multiple cloning sites. Gene. 1987;51(2-3):275–280. doi: 10.1016/0378-1119(87)90316-7. [DOI] [PubMed] [Google Scholar]
- Doten R. C., Gregg L. A., Ornston L. N. Influence of the catBCE sequence on the phenotypic reversion of a pcaE mutation in Acinetobacter calcoaceticus. J Bacteriol. 1987 Jul;169(7):3175–3180. doi: 10.1128/jb.169.7.3175-3180.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durham D. R., Phibbs P. V., Jr Fractionation and characterization of the phosphoenolpyruvate: fructose 1-phosphotransferase system from Pseudomonas aeruginosa. J Bacteriol. 1982 Feb;149(2):534–541. doi: 10.1128/jb.149.2.534-541.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensley B. D., Gibson D. T., Laborde A. L. Oxidation of naphthalene by a multicomponent enzyme system from Pseudomonas sp. strain NCIB 9816. J Bacteriol. 1982 Mar;149(3):948–954. doi: 10.1128/jb.149.3.948-954.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensley B. D., Gibson D. T. Naphthalene dioxygenase: purification and properties of a terminal oxygenase component. J Bacteriol. 1983 Aug;155(2):505–511. doi: 10.1128/jb.155.2.505-511.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin F. C., Bagdasarian M., Bagdasarian M. M., Timmis K. N. Molecular and functional analysis of the TOL plasmid pWWO from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta cleavage pathway. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7458–7462. doi: 10.1073/pnas.78.12.7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürste J. P., Pansegrau W., Frank R., Blöcker H., Scholz P., Bagdasarian M., Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48(1):119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- Gibson D. T., Hensley M., Yoshioka H., Mabry T. J. Formation of (+)-cis-2,3-dihydroxy-1-methylcyclohexa-4,6-diene from toluene by Pseudomonas putida. Biochemistry. 1970 Mar 31;9(7):1626–1630. doi: 10.1021/bi00809a023. [DOI] [PubMed] [Google Scholar]
- Gibson D. T., Roberts R. L., Wells M. C., Kobal V. M. Oxidation of biphenyl by a Beijerinckia species. Biochem Biophys Res Commun. 1973 Jan 23;50(2):211–219. doi: 10.1016/0006-291x(73)90828-0. [DOI] [PubMed] [Google Scholar]
- HUGHES D. E. A press for disrupting bacteria and other micro-organisms. Br J Exp Pathol. 1951 Apr;32(2):97–109. [PMC free article] [PubMed] [Google Scholar]
- Harayama S., Lehrbach P. R., Timmis K. N. Transposon mutagenesis analysis of meta-cleavage pathway operon genes of the TOL plasmid of Pseudomonas putida mt-2. J Bacteriol. 1984 Oct;160(1):251–255. doi: 10.1128/jb.160.1.251-255.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harayama S., Rekik M. The meta cleavage operon of TOL degradative plasmid pWW0 comprises 13 genes. Mol Gen Genet. 1990 Mar;221(1):113–120. doi: 10.1007/BF00280375. [DOI] [PubMed] [Google Scholar]
- Harayama S., Rekik M., Timmis K. N. Genetic analysis of a relaxed substrate specificity aromatic ring dioxygenase, toluate 1,2-dioxygenase, encoded by TOL plasmid pWW0 of Pseudomonas putida. Mol Gen Genet. 1986 Feb;202(2):226–234. doi: 10.1007/BF00331641. [DOI] [PubMed] [Google Scholar]
- Hareland W. A., Crawford R. L., Chapman P. J., Dagley S. Metabolic function and properties of 4-hydroxyphenylacetic acid 1-hydroxylase from Pseudomonas acidovorans. J Bacteriol. 1975 Jan;121(1):272–285. doi: 10.1128/jb.121.1.272-285.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugouvieux-Cotte-Pattat N., Köhler T., Rekik M., Harayama S. Growth-phase-dependent expression of the Pseudomonas putida TOL plasmid pWW0 catabolic genes. J Bacteriol. 1990 Dec;172(12):6651–6660. doi: 10.1128/jb.172.12.6651-6660.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hylemon P. B., Phibbs P. V., Jr Independent regulation of hexose catabolizing enzymes and glucose transport activity in Pseudomonas aeruginosa. Biochem Biophys Res Commun. 1972 Sep 5;48(5):1041–1048. doi: 10.1016/0006-291x(72)90813-3. [DOI] [PubMed] [Google Scholar]
- ICHIHARA A., ADACHI K., HOSOKAWA K., TAKEDA Y. The enzymatic hydroxylation of aromatic carboxylic acids; substrate specificities of anthranilate and benzoate oxidases. J Biol Chem. 1962 Jul;237:2296–2302. [PubMed] [Google Scholar]
- Inouye S., Nakazawa A., Nakazawa T. Molecular cloning of gene xylS of the TOL plasmid: evidence for positive regulation of the xylDEGF operon by xylS. J Bacteriol. 1981 Nov;148(2):413–418. doi: 10.1128/jb.148.2.413-418.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie S., Doi S., Yorifuji T., Takagi M., Yano K. Nucleotide sequencing and characterization of the genes encoding benzene oxidation enzymes of Pseudomonas putida. J Bacteriol. 1987 Nov;169(11):5174–5179. doi: 10.1128/jb.169.11.5174-5179.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukor J. J., Olsen R. H., Ballou D. P. Cloning and expression of the catA and catBC gene clusters from Pseudomonas aeruginosa PAO. J Bacteriol. 1988 Oct;170(10):4458–4465. doi: 10.1128/jb.170.10.4458-4465.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurkela S., Lehväslaiho H., Palva E. T., Teeri T. H. Cloning, nucleotide sequence and characterization of genes encoding naphthalene dioxygenase of Pseudomonas putida strain NCIB9816. Gene. 1988 Dec 20;73(2):355–362. doi: 10.1016/0378-1119(88)90500-8. [DOI] [PubMed] [Google Scholar]
- Mermod N., Ramos J. L., Bairoch A., Timmis K. N. The xylS gene positive regulator of TOL plasmid pWWO: identification, sequence analysis and overproduction leading to constitutive expression of meta cleavage operon. Mol Gen Genet. 1987 May;207(2-3):349–354. doi: 10.1007/BF00331600. [DOI] [PubMed] [Google Scholar]
- Neidle E. L., Hartnett C., Bonitz S., Ornston L. N. DNA sequence of the Acinetobacter calcoaceticus catechol 1,2-dioxygenase I structural gene catA: evidence for evolutionary divergence of intradiol dioxygenases by acquisition of DNA sequence repetitions. J Bacteriol. 1988 Oct;170(10):4874–4880. doi: 10.1128/jb.170.10.4874-4880.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidle E. L., Hartnett C., Ornston L. N., Bairoch A., Rekik M., Harayama S. Nucleotide sequences of the Acinetobacter calcoaceticus benABC genes for benzoate 1,2-dioxygenase reveal evolutionary relationships among multicomponent oxygenases. J Bacteriol. 1991 Sep;173(17):5385–5395. doi: 10.1128/jb.173.17.5385-5395.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidle E. L., Shapiro M. K., Ornston L. N. Cloning and expression in Escherichia coli of Acinetobacter calcoaceticus genes for benzoate degradation. J Bacteriol. 1987 Dec;169(12):5496–5503. doi: 10.1128/jb.169.12.5496-5503.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen R. H., DeBusscher G., McCombie W. R. Development of broad-host-range vectors and gene banks: self-cloning of the Pseudomonas aeruginosa PAO chromosome. J Bacteriol. 1982 Apr;150(1):60–69. doi: 10.1128/jb.150.1.60-69.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen R. H., Shipley P. Host range and properties of the Pseudomonas aeruginosa R factor R1822. J Bacteriol. 1973 Feb;113(2):772–780. doi: 10.1128/jb.113.2.772-780.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornston L. N. Regulation of catabolic pathways in Pseudomonas. Bacteriol Rev. 1971 Jun;35(2):87–116. doi: 10.1128/br.35.2.87-116.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornston L. N., Stanier R. Y. The conversion of catechol and protocatechuate to beta-ketoadipate by Pseudomonas putida. J Biol Chem. 1966 Aug 25;241(16):3776–3786. [PubMed] [Google Scholar]
- Ornston L. N. The conversion of catechol and protocatechuate to beta-ketoadipate by Pseudomonas putida. 3. Enzymes of the catechol pathway. J Biol Chem. 1966 Aug 25;241(16):3795–3799. [PubMed] [Google Scholar]
- Ornston L. N. The conversion of catechol and protocatechuate to beta-ketoadipate by Pseudomonas putida. II. Enzymes of the protocatechuate pathway. J Biol Chem. 1966 Aug 25;241(16):3787–3794. [PubMed] [Google Scholar]
- Ramos J. L., Mermod N., Timmis K. N. Regulatory circuits controlling transcription of TOL plasmid operon encoding meta-cleavage pathway for degradation of alkylbenzoates by Pseudomonas. Mol Microbiol. 1987 Nov;1(3):293–300. doi: 10.1111/j.1365-2958.1987.tb01935.x. [DOI] [PubMed] [Google Scholar]
- Reiner A. M., Hegeman G. D. Metabolism of benzoic acid by bacteria. Accumulation of (-)-3,5-cyclohexadiene-1,2-diol-1-carboxylic acid by mutant strain of Alcaligenes eutrophus. Biochemistry. 1971 Jun 22;10(13):2530–2536. doi: 10.1021/bi00789a017. [DOI] [PubMed] [Google Scholar]
- Reiner A. M. Metabolism of benzoic acid by bacteria: 3,5-cyclohexadiene-1,2-diol-1-carboxylic acid is an intermediate in the formation of catechol. J Bacteriol. 1971 Oct;108(1):89–94. doi: 10.1128/jb.108.1.89-94.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roehl R. A., Phibbs P. V., Jr Characterization and genetic mapping of fructose phosphotransferase mutations in Pseudomonas aeruginosa. J Bacteriol. 1982 Mar;149(3):897–905. doi: 10.1128/jb.149.3.897-905.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg S. L., Hegeman G. D. Clustering of functionally related genes in Pseudomonas aeruginosa. J Bacteriol. 1969 Jul;99(1):353–355. doi: 10.1128/jb.99.1.353-355.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanley M. S., Neidle E. L., Parales R. E., Ornston L. N. Cloning and expression of Acinetobacter calcoaceticus catBCDE genes in Pseudomonas putida and Escherichia coli. J Bacteriol. 1986 Feb;165(2):557–563. doi: 10.1128/jb.165.2.557-563.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields M. S., Montgomery S. O., Chapman P. J., Cuskey S. M., Pritchard P. H. Novel pathway of toluene catabolism in the trichloroethylene-degrading bacterium g4. Appl Environ Microbiol. 1989 Jun;55(6):1624–1629. doi: 10.1128/aem.55.6.1624-1629.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985 Oct;150(1):76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Wheelis M. L., Stanier R. Y. The genetic control of dissimilatory pathways in Pseudomonas putida. Genetics. 1970 Oct;66(2):245–266. doi: 10.1093/genetics/66.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M., Fujisawa H. Characterization of NADH-cytochrome c reductase, a component of benzoate 1,2-dioxygenase system from Pseudomonas arvilla c-1. J Biol Chem. 1978 Dec 25;253(24):8848–8853. [PubMed] [Google Scholar]
- Yamaguchi M., Fujisawa H. Purification and characterization of an oxygenase component in benzoate 1,2-dioxygenase system from Pseudomonas arvilla C-1. J Biol Chem. 1980 Jun 10;255(11):5058–5063. [PubMed] [Google Scholar]
- Yamaguchi M., Fujisawa H. Subunit structure of oxygenase component in benzoate-1,2-dioxygenase system from Pseudomonas arvilla C-1. J Biol Chem. 1982 Nov 10;257(21):12497–12502. [PubMed] [Google Scholar]
- Zeyer J., Lehrbach P. R., Timmis K. N. Use of cloned genes of Pseudomonas TOL plasmid to effect biotransformation of benzoates to cis-dihydrodiols and catechols by Escherichia coli cells. Appl Environ Microbiol. 1985 Dec;50(6):1409–1413. doi: 10.1128/aem.50.6.1409-1413.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L. M., Timmis K. N., Ramos J. L. Mutations leading to constitutive expression from the TOL plasmid meta-cleavage pathway operon are located at the C-terminal end of the positive regulator protein XylS. J Bacteriol. 1990 Jul;172(7):3707–3710. doi: 10.1128/jb.172.7.3707-3710.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylstra G. J., Gibson D. T. Toluene degradation by Pseudomonas putida F1. Nucleotide sequence of the todC1C2BADE genes and their expression in Escherichia coli. J Biol Chem. 1989 Sep 5;264(25):14940–14946. [PubMed] [Google Scholar]



