Abstract
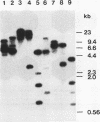
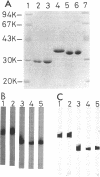
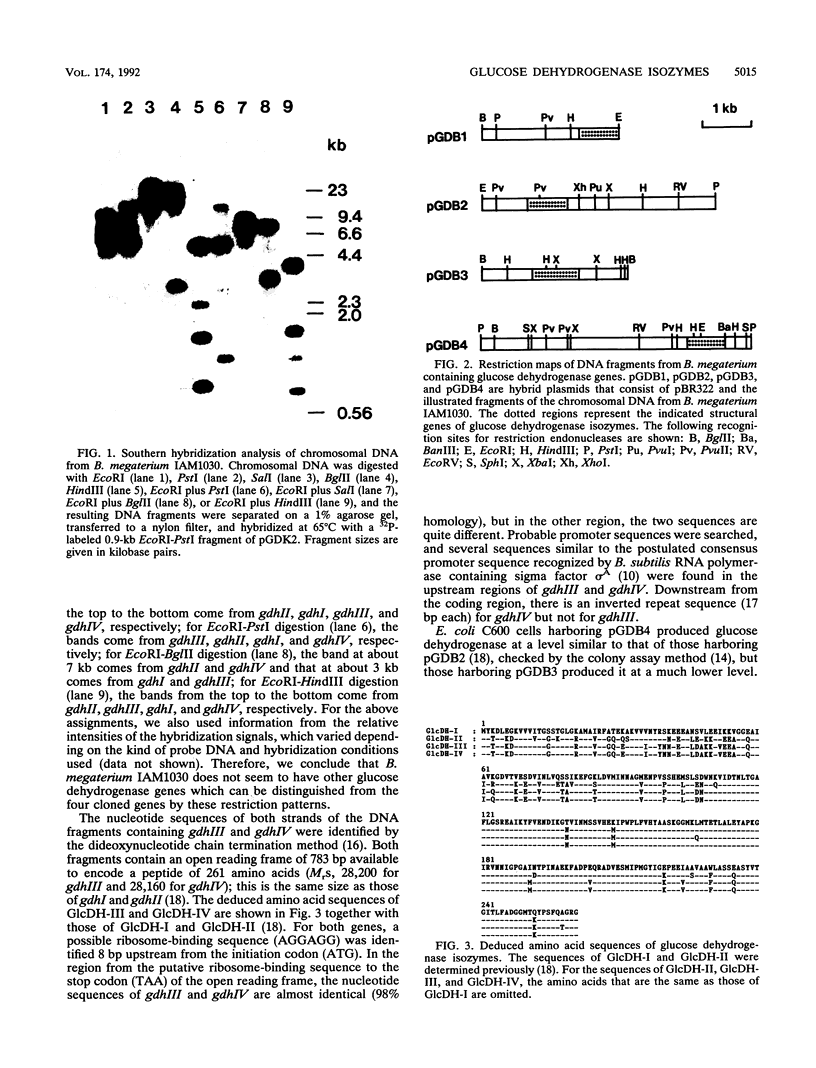
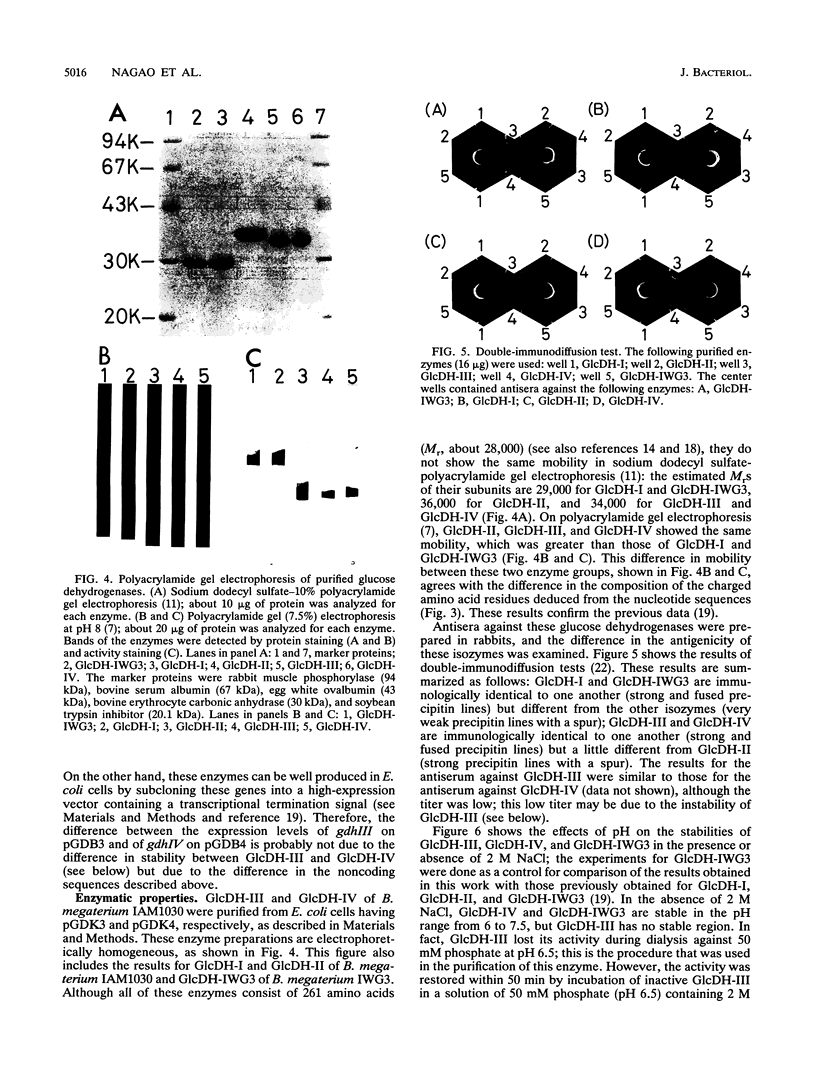
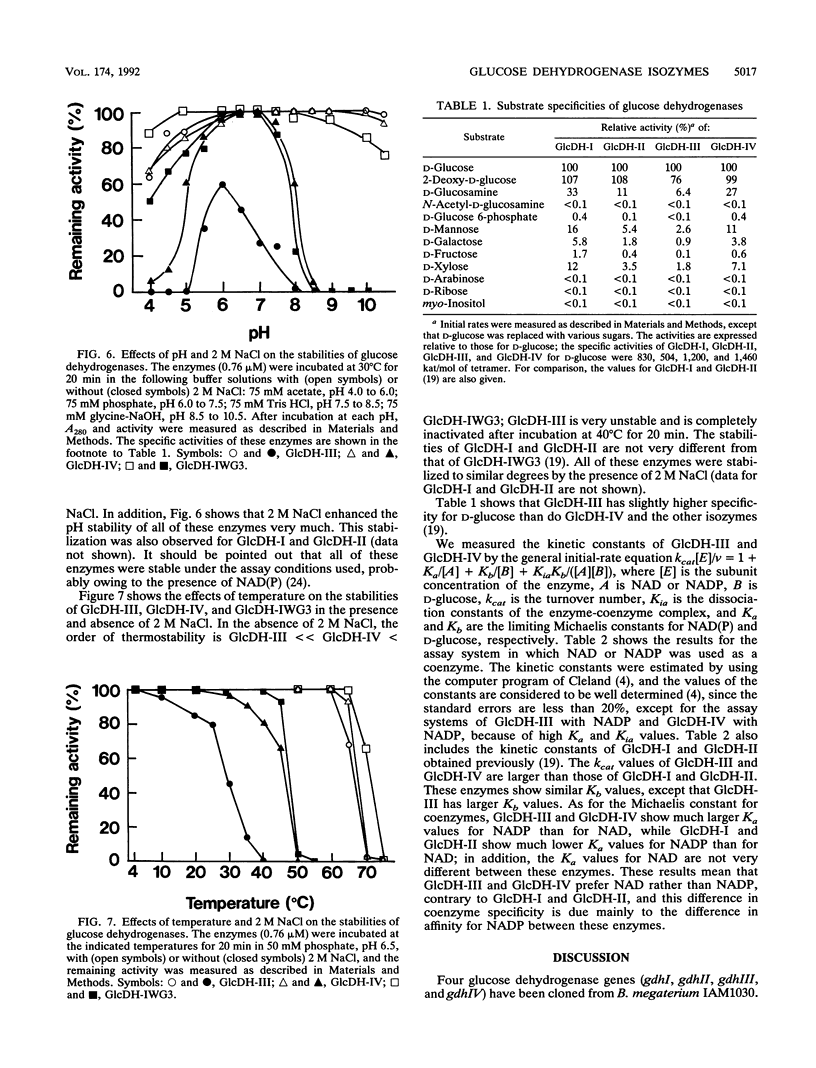
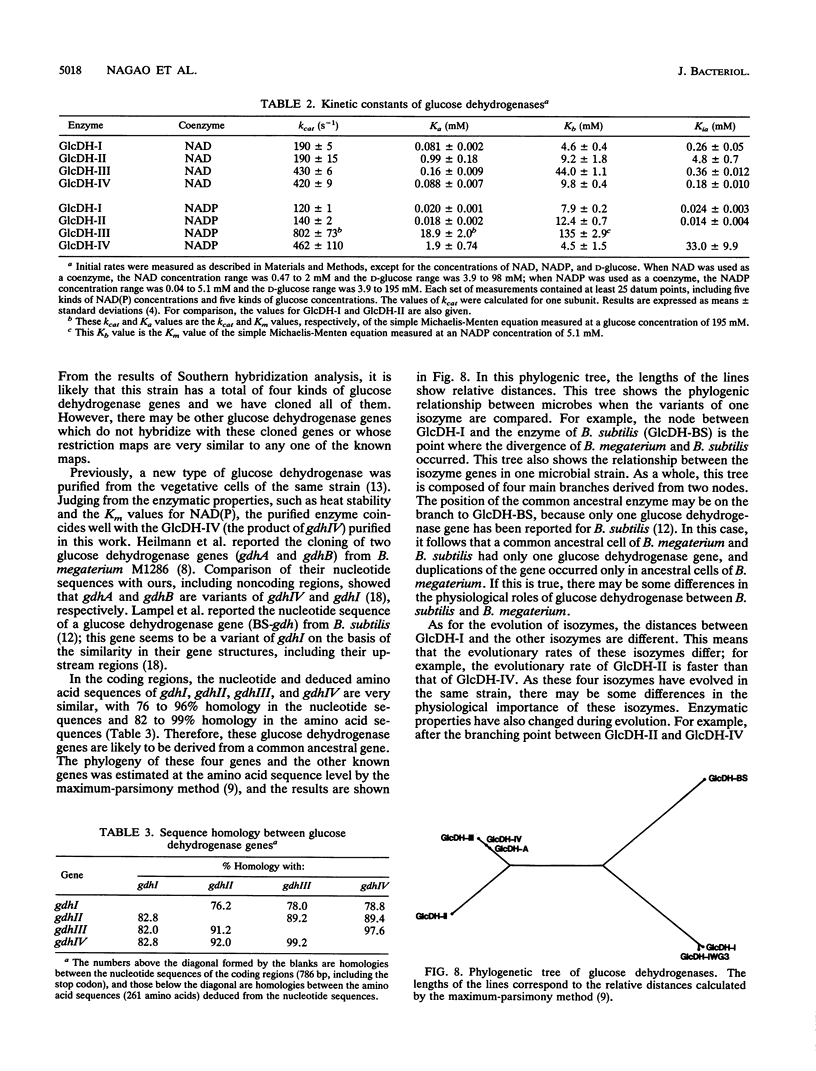
Bacillus megaterium is known to have several genes that code for isozymes of glucose dehydrogenase. Two of them, gdhI and gdhII, were cloned from B. megaterium IAM1030 in our previous work (T. Mitamura, R. V. Evora, T. Nakai, Y. Makino, S. Negoro, I. Urabe, and H. Okada, J. Ferment. Bioeng. 70:363-369, 1990). In the present study, two new genes, gdhIII and gdhIV, were isolated from the same strain and their nucleotide sequences were identified. Each gene has an open reading frame of 783 bp available to encode a peptide of 261 amino acids. Thus, a total of four glucose dehydrogenase genes have been cloned from B. megaterium IAM1030. In addition, this strain does not seem to have other glucose dehydrogenase genes that can be distinguished from the four cloned genes so far examined by Southern hybridization analysis. The two newly cloned genes were expressed in Escherichia coli cells, and the products, GlcDH-III and GlcDH-IV, were purified and characterized and compared with the other isozymes, GlcDH-I and GlcDH-II, encoded by gdhI and gdhII, respectively. These isozymes showed different mobilities in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (GlcDH-I greater than GlcDH-III = GlcDH-IV greater than GlcDH-II), although they have the same number of amino acid residues. Double-immunodiffusion tests showed that GlcDH-I is immunologically different from the other isozymes and that GlcDH-III and GlcDH-IV are identical to one another but a little different from GlcDH-II. These glucose dehydrogenases were stabilized in the presence of 2 M NaCl. The effect of NaCl was especially large for GlcDH-III, which is most unstable enzyme. Kinetic studies showed that these isozymes are divided into two groups with respect to coenzyme specificity, although they can utilize both NAD and NADP: GlcDH-III and GlcDH-IV prefer NAD, and GlcDH-I and GlcDH-II prefer NADP. The phylogenic relationship of these glucose dehydrogenase genes is also discussed.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BACH J. A., SADOFF H. L. Aerobic sporulating bacteria. I. Glucose dehydrogenase of Bacillus cereus. J Bacteriol. 1962 Apr;83:699–707. doi: 10.1128/jb.83.4.699-707.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972 Dec;36(4):525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Cleland W. W. Statistical analysis of enzyme kinetic data. Methods Enzymol. 1979;63:103–138. doi: 10.1016/0076-6879(79)63008-2. [DOI] [PubMed] [Google Scholar]
- Fujita Y., Ramaley R., Freese E. Location and properties of glucose dehydrogenase in sporulating cells and spores of Bacillus subtilis. J Bacteriol. 1977 Oct;132(1):282–293. doi: 10.1128/jb.132.1.282-293.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann H. J., Mägert H. J., Gassen H. G. Identification and isolation of glucose dehydrogenase genes of Bacillus megaterium M1286 and their expression in Escherichia coli. Eur J Biochem. 1988 Jun 15;174(3):485–490. doi: 10.1111/j.1432-1033.1988.tb14124.x. [DOI] [PubMed] [Google Scholar]
- Hein J. Unified approach to alignment and phylogenies. Methods Enzymol. 1990;183:626–645. doi: 10.1016/0076-6879(90)83041-7. [DOI] [PubMed] [Google Scholar]
- Helmann J. D., Chamberlin M. J. Structure and function of bacterial sigma factors. Annu Rev Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lampel K. A., Uratani B., Chaudhry G. R., Ramaley R. F., Rudikoff S. Characterization of the developmentally regulated Bacillus subtilis glucose dehydrogenase gene. J Bacteriol. 1986 Apr;166(1):238–243. doi: 10.1128/jb.166.1.238-243.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino Y., Negoro S., Urabe I., Okada H. Stability-increasing mutants of glucose dehydrogenase from Bacillus megaterium IWG3. J Biol Chem. 1989 Apr 15;264(11):6381–6385. [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T., Yasukochi T., Nagatani H., Furuno M., Orita T., Yamada H., Imoto T., Horiuchi T. Construction of a plasmid vector for the regulatable high level expression of eukaryotic genes in Escherichia coli: an application to overproduction of chicken lysozyme. Protein Eng. 1987 Aug-Sep;1(4):327–332. doi: 10.1093/protein/1.4.327. [DOI] [PubMed] [Google Scholar]
- Mitamura T., Urabe I., Okada H. Enzymatic properties of isozymes and variants of glucose dehydrogenase from Bacillus megaterium. Eur J Biochem. 1989 Dec 8;186(1-2):389–393. doi: 10.1111/j.1432-1033.1989.tb15221.x. [DOI] [PubMed] [Google Scholar]
- Nagao T., Makino Y., Yamamoto K., Urabe I., Okada H. Stability-increasing mutants of glucose dehydrogenase. FEBS Lett. 1989 Aug 14;253(1-2):113–116. doi: 10.1016/0014-5793(89)80941-x. [DOI] [PubMed] [Google Scholar]
- Nakatani Y., Nicholson W. L., Neitzke K. D., Setlow P., Freese E. Sigma-G RNA polymerase controls forespore-specific expression of the glucose dehydrogenase operon in Bacillus subtilis. Nucleic Acids Res. 1989 Feb 11;17(3):999–1017. doi: 10.1093/nar/17.3.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauly H. E., Pfleiderer G. D-glucose dehydrogenase from Bacillus megaterium M 1286: purification, properties and structure. Hoppe Seylers Z Physiol Chem. 1975 Oct;356(10):1613–1623. doi: 10.1515/bchm2.1975.356.2.1613. [DOI] [PubMed] [Google Scholar]
- SAITO H., MIURA K. I. PREPARATION OF TRANSFORMING DEOXYRIBONUCLEIC ACID BY PHENOL TREATMENT. Biochim Biophys Acta. 1963 Aug 20;72:619–629. [PubMed] [Google Scholar]
- Sano K., Otani M., Umezawa C. Glucose metabolism via the Embden-Meyerhof pathway is not involved in ATP production during spore germination of bacillus megaterium QM B1551. A study with a mutant lacking hexokinase. Biochem Biophys Res Commun. 1988 Feb 29;151(1):48–52. doi: 10.1016/0006-291x(88)90557-8. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Vogelstein B., Gillespie D. Preparative and analytical purification of DNA from agarose. Proc Natl Acad Sci U S A. 1979 Feb;76(2):615–619. doi: 10.1073/pnas.76.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]