Abstract
A molecular ecological study was performed on an Australian soil sample to unravel a substantial portion of the bacterial diversity. A large fragment of the 16S rRNA gene was amplified, using DNA isolated by lysing the microorganisms directly within the soil matrix, and a clone library was generated. Comparative sequence analysis of 30 clones and dot blot hybridization of 83 additional clones with defined oligonucleotide probes revealed the presence of three major groups of prokaryotes of the domain Bacteria. The first one comprises 57 clones that indicate relatives of nitrogen-fixing bacteria of the alpha-2 subclass of the class Proteobacteria; the second group of 7 clones originates from members of the order Planctomycetales that, however, reveal no close relationship to any of the described Planctomycetales species; 22 clones of the third group are indicative of members of a novel main line of descent, sharing a common ancestry with members of planctomycetes and chlamydiae.
Full text
PDF

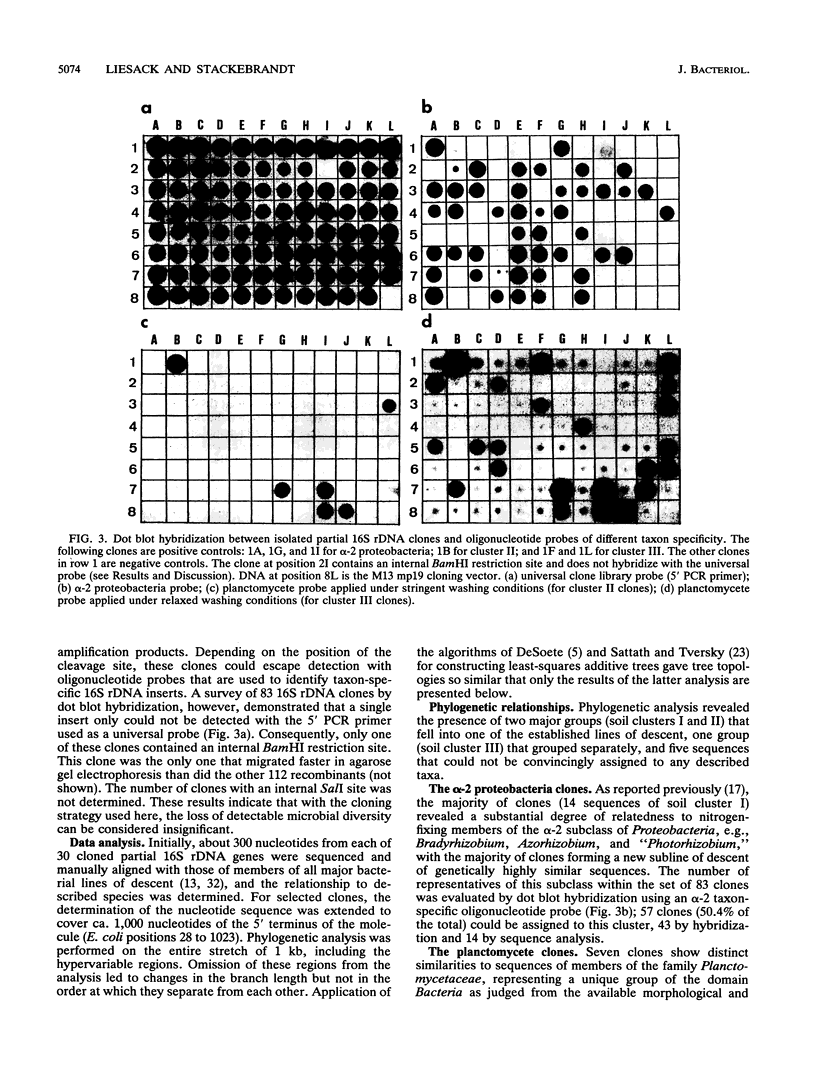




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amann R., Springer N., Ludwig W., Görtz H. D., Schleifer K. H. Identification in situ and phylogeny of uncultured bacterial endosymbionts. Nature. 1991 May 9;351(6322):161–164. doi: 10.1038/351161a0. [DOI] [PubMed] [Google Scholar]
- Bakken L. R. Separation and purification of bacteria from soil. Appl Environ Microbiol. 1985 Jun;49(6):1482–1487. doi: 10.1128/aem.49.6.1482-1487.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britschgi T. B., Giovannoni S. J. Phylogenetic analysis of a natural marine bacterioplankton population by rRNA gene cloning and sequencing. Appl Environ Microbiol. 1991 Jun;57(6):1707–1713. doi: 10.1128/aem.57.6.1707-1713.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzmann P. D., Skerman V. B. Gemmata obscuriglobus, a new genus and species of the budding bacteria. Antonie Van Leeuwenhoek. 1984;50(3):261–268. doi: 10.1007/BF02342136. [DOI] [PubMed] [Google Scholar]
- Fuerst J. A., Webb R. I. Membrane-bounded nucleoid in the eubacterium Gemmata obscuriglobus. Proc Natl Acad Sci U S A. 1991 Sep 15;88(18):8184–8188. doi: 10.1073/pnas.88.18.8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni S. J., Britschgi T. B., Moyer C. L., Field K. G. Genetic diversity in Sargasso Sea bacterioplankton. Nature. 1990 May 3;345(6270):60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- Holben William E., Jansson Janet K., Chelm Barry K., Tiedje James M. DNA Probe Method for the Detection of Specific Microorganisms in the Soil Bacterial Community. Appl Environ Microbiol. 1988 Mar;54(3):703–711. doi: 10.1128/aem.54.3.703-711.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesack W., Stackebrandt E. Evidence for unlinked rrn operons in the Planctomycete Pirellula marina. J Bacteriol. 1989 Sep;171(9):5025–5030. doi: 10.1128/jb.171.9.5025-5030.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neefs J. M., Van de Peer Y., Hendriks L., De Wachter R. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 1990 Apr 25;18 (Suppl):2237–2317. doi: 10.1093/nar/18.suppl.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen G. J., Lane D. J., Giovannoni S. J., Pace N. R., Stahl D. A. Microbial ecology and evolution: a ribosomal RNA approach. Annu Rev Microbiol. 1986;40:337–365. doi: 10.1146/annurev.mi.40.100186.002005. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Stackebrandt E., Charfreitag O. Partial 16S rRNA primary structure of five Actinomyces species: phylogenetic implications and development of an Actinomyces israelii-specific oligonucleotide probe. J Gen Microbiol. 1990 Jan;136(1):37–43. doi: 10.1099/00221287-136-1-37. [DOI] [PubMed] [Google Scholar]
- Steffan R. J., Atlas R. M. Polymerase chain reaction: applications in environmental microbiology. Annu Rev Microbiol. 1991;45:137–161. doi: 10.1146/annurev.mi.45.100191.001033. [DOI] [PubMed] [Google Scholar]
- Steffan R. J., Goksøyr J., Bej A. K., Atlas R. M. Recovery of DNA from soils and sediments. Appl Environ Microbiol. 1988 Dec;54(12):2908–2915. doi: 10.1128/aem.54.12.2908-2915.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward D. M., Weller R., Bateson M. M. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature. 1990 May 3;345(6270):63–65. doi: 10.1038/345063a0. [DOI] [PubMed] [Google Scholar]
- Weisburg W. G., Barns S. M., Pelletier D. A., Lane D. J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991 Jan;173(2):697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisburg W. G., Hatch T. P., Woese C. R. Eubacterial origin of chlamydiae. J Bacteriol. 1986 Aug;167(2):570–574. doi: 10.1128/jb.167.2.570-574.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller R., Weller J. W., Ward D. M. 16S rRNA sequences of uncultivated hot spring cyanobacterial mat inhabitants retrieved as randomly primed cDNA. Appl Environ Microbiol. 1991 Apr;57(4):1146–1151. doi: 10.1128/aem.57.4.1146-1151.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]





