Abstract
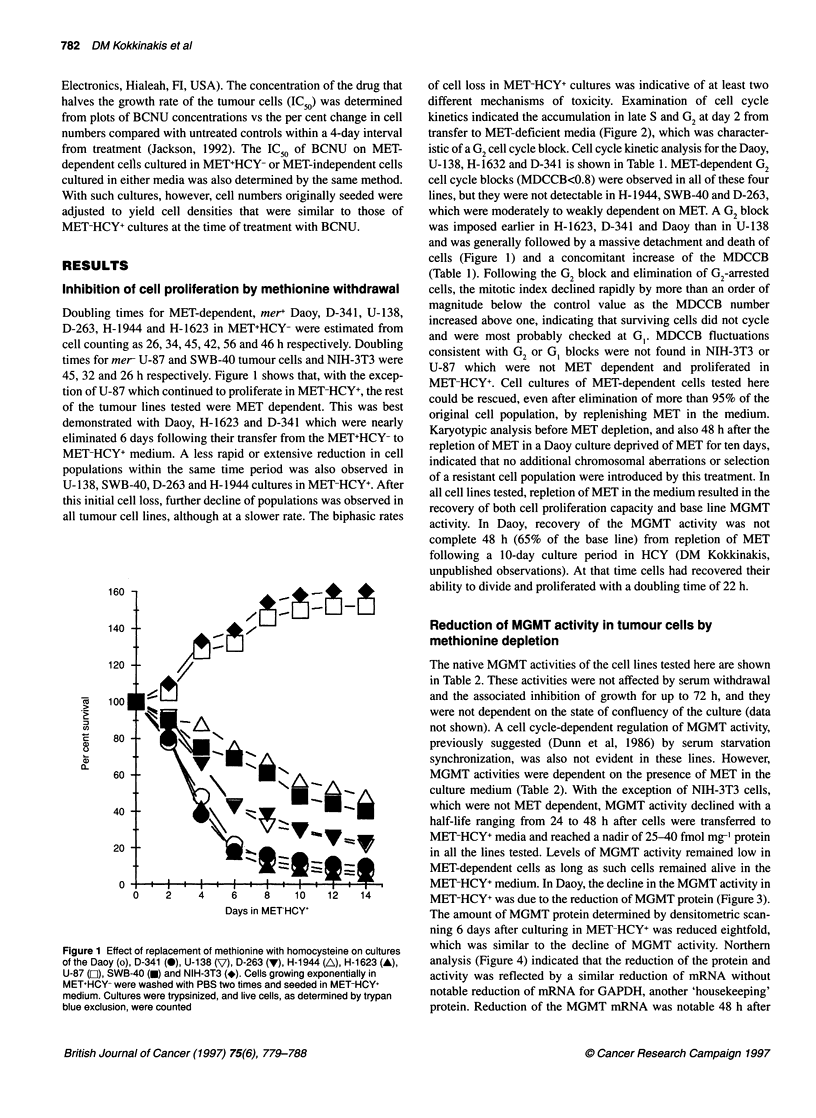
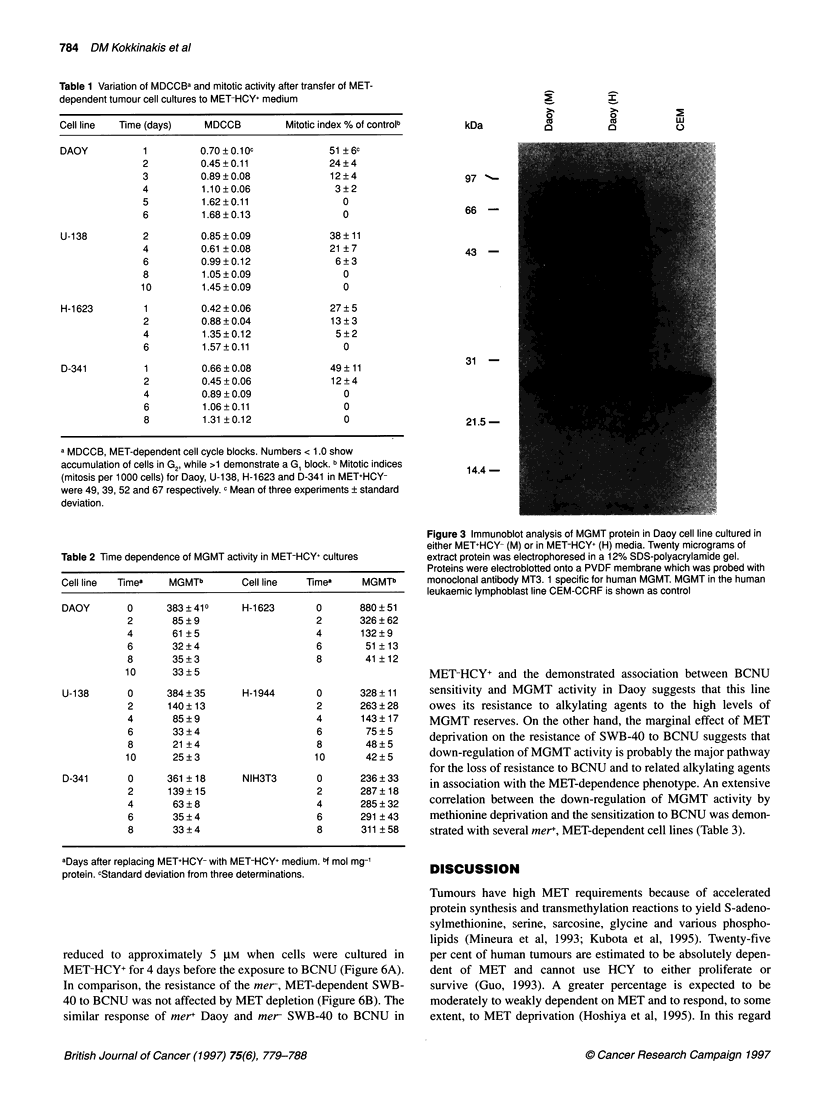
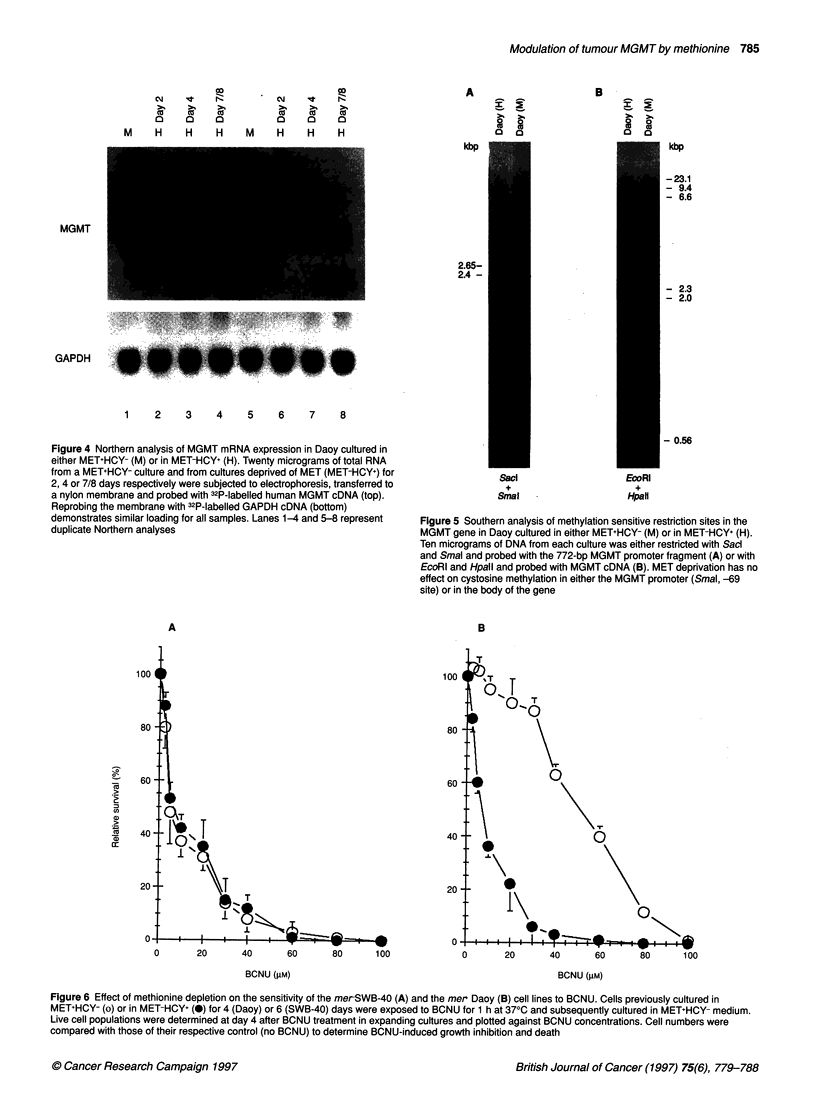
Methionine (MET)-dependent cell lines require MET to proliferate, and homocysteine (HCY) does not act as a substitute for this requirement. From six O6-methylguanine-DNA methyltransferase (MGMT)-efficient (mer+) cell lines tested, two medulloblastomas (Daoy and D-341) and a lung non-small-cell adenocarcinoma with metastatic potential (H-1623) were most sensitive to MET deprivation, while two glioblastomas (U-138, D-263) and a small-cell lung carcinoma H-1944 were moderately to weakly dependent. Regardless of the degree of MET dependence, all of these lines down-regulated their MGMT activity within 48-72 h of transfer from MET+HCY- to MET-HCY+ media, long before the eradication of the culture. Reduction of MGMT activity was due to a decline of both MGMT mRNA and protein levels. However, the reduction was not related to the methylation status of the MGMT promoter at the SmaI site or the HpaII sites in the body of the gene; such sites have been shown to be associated in MGMT regulation and in defining the mer phenotype. MET-dependent, mer+ tumour cells cultured in MET-HCY+ were more sensitive to BCNU (IC50 = 5-10 microM) than those cultured in MET+HCY-(IC50 = 45-90 microM), while MET-independent or mer- cell lines were unaffected. This indicates that reduction of MGMT, imposed by the absence of MET, renders mer+ tumour cells more susceptible to alkylating agents. The relatively selective suppression of MGMT activity in mer+ MET-dependent tumour cells, in combination with the inability of such cells to proliferate in the absence of MET, may lead to the development of more effective treatment strategies for mer+ MET-dependent tumours.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Branch P., Aquilina G., Bignami M., Karran P. Defective mismatch binding and a mutator phenotype in cells tolerant to DNA damage. Nature. 1993 Apr 15;362(6421):652–654. doi: 10.1038/362652a0. [DOI] [PubMed] [Google Scholar]
- Breillout F., Antoine E., Poupon M. F. Methionine dependency of malignant tumors: a possible approach for therapy. J Natl Cancer Inst. 1990 Oct 17;82(20):1628–1632. doi: 10.1093/jnci/82.20.1628. [DOI] [PubMed] [Google Scholar]
- Breillout F., Hadida F., Echinard-Garin P., Lascaux V., Poupon M. F. Decreased rat rhabdomyosarcoma pulmonary metastases in response to a low methionine diet. Anticancer Res. 1987 Jul-Aug;7(4B):861–867. [PubMed] [Google Scholar]
- Brent T. P., von Wronski M. A., Edwards C. C., Bromley M., Margison G. P., Rafferty J. A., Pegram C. N., Bigner D. D. Identification of nitrosourea-resistant human rhabdomyosarcomas by in situ immunostaining of O6-methylguanine-DNA methyltransferase. Oncol Res. 1993;5(2):83–86. [PubMed] [Google Scholar]
- Costello J. F., Futscher B. W., Kroes R. A., Pieper R. O. Methylation-related chromatin structure is associated with exclusion of transcription factors from and suppressed expression of the O-6-methylguanine DNA methyltransferase gene in human glioma cell lines. Mol Cell Biol. 1994 Oct;14(10):6515–6521. doi: 10.1128/mcb.14.10.6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day R. S., 3rd, Ziolkowski C. H., Scudiero D. A., Meyer S. A., Lubiniecki A. S., Girardi A. J., Galloway S. M., Bynum G. D. Defective repair of alkylated DNA by human tumour and SV40-transformed human cell strains. Nature. 1980 Dec 25;288(5792):724–727. doi: 10.1038/288724a0. [DOI] [PubMed] [Google Scholar]
- Dolan M. E., Pegg A. E., Moschel R. C., Grindey G. B. Effect of O6-benzylguanine on the sensitivity of human colon tumor xenografts to 1,3-bis(2-chloroethyl)-1-nitrosourea (BCNU). Biochem Pharmacol. 1993 Jul 20;46(2):285–290. doi: 10.1016/0006-2952(93)90416-t. [DOI] [PubMed] [Google Scholar]
- Dolan M. E., Stine L., Mitchell R. B., Moschel R. C., Pegg A. E. Modulation of mammalian O6-alkylguanine-DNA alkyltransferase in vivo by O6-benzylguanine and its effect on the sensitivity of a human glioma tumor to 1-(2-chloroethyl)-3-(4-methylcyclohexyl)-1-nitrosourea. Cancer Commun. 1990;2(11):371–377. doi: 10.3727/095535490820873985. [DOI] [PubMed] [Google Scholar]
- Dunn W. C., Foote R. S., Hand R. E., Jr, Mitra S. Cell cycle-dependent modulation of O6-methylguanine-DNA methyltransferase in C3H/10T1/2 cells. Carcinogenesis. 1986 May;7(5):807–812. doi: 10.1093/carcin/7.5.807. [DOI] [PubMed] [Google Scholar]
- Dunn W. C., Tano K., Horesovsky G. J., Preston R. J., Mitra S. The role of O6-alkylguanine in cell killing and mutagenesis in Chinese hamster ovary cells. Carcinogenesis. 1991 Jan;12(1):83–89. doi: 10.1093/carcin/12.1.83. [DOI] [PubMed] [Google Scholar]
- Egyházi S., Bergh J., Hansson J., Karran P., Ringborg U. Carmustine-induced toxicity, DNA crosslinking and O6-methylguanine-DNA methyltransferase activity in two human lung cancer cell lines. Eur J Cancer. 1991;27(12):1658–1662. doi: 10.1016/0277-5379(91)90440-o. [DOI] [PubMed] [Google Scholar]
- Finkelstein J. D. Methionine metabolism in mammals. J Nutr Biochem. 1990 May;1(5):228–237. doi: 10.1016/0955-2863(90)90070-2. [DOI] [PubMed] [Google Scholar]
- Fiskerstrand T., Christensen B., Tysnes O. B., Ueland P. M., Refsum H. Development and reversion of methionine dependence in a human glioma cell line: relation to homocysteine remethylation and cobalamin status. Cancer Res. 1994 Sep 15;54(18):4899–4906. [PubMed] [Google Scholar]
- Gerson S. L., Miller K., Berger N. A. O6 alkylguanine-DNA alkyltransferase activity in human myeloid cells. J Clin Invest. 1985 Dec;76(6):2106–2114. doi: 10.1172/JCI112215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goseki N., Yamazaki S., Endo M., Onodera T., Kosaki G., Hibino Y., Kuwahata T. Antitumor effect of methionine-depleting total parenteral nutrition with doxorubicin administration on Yoshida sarcoma-bearing rats. Cancer. 1992 Apr 1;69(7):1865–1872. doi: 10.1002/1097-0142(19920401)69:7<1865::aid-cncr2820690732>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Guo H. Y., Herrera H., Groce A., Hoffman R. M. Expression of the biochemical defect of methionine dependence in fresh patient tumors in primary histoculture. Cancer Res. 1993 Jun 1;53(11):2479–2483. [PubMed] [Google Scholar]
- Guo H., Lishko V. K., Herrera H., Groce A., Kubota T., Hoffman R. M. Therapeutic tumor-specific cell cycle block induced by methionine starvation in vivo. Cancer Res. 1993 Dec 1;53(23):5676–5679. [PubMed] [Google Scholar]
- Halpern B. C., Clark B. R., Hardy D. N., Halpern R. M., Smith R. A. The effect of replacement of methionine by homocystine on survival of malignant and normal adult mammalian cells in culture. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1133–1136. doi: 10.1073/pnas.71.4.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris L. C., Potter P. M., Tano K., Shiota S., Mitra S., Brent T. P. Characterization of the promoter region of the human O6-methylguanine-DNA methyltransferase gene. Nucleic Acids Res. 1991 Nov 25;19(22):6163–6167. doi: 10.1093/nar/19.22.6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris L. C., Remack J. S., Brent T. P. In vitro methylation of the human O6-methylguanine-DNA methyltransferase promoter reduces transcription. Biochim Biophys Acta. 1994 Mar 1;1217(2):141–146. doi: 10.1016/0167-4781(94)90027-2. [DOI] [PubMed] [Google Scholar]
- Hoffman R. M., Erbe R. W. High in vivo rates of methionine biosynthesis in transformed human and malignant rat cells auxotrophic for methionine. Proc Natl Acad Sci U S A. 1976 May;73(5):1523–1527. doi: 10.1073/pnas.73.5.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman R. M. Unbalanced transmethylation and the perturbation of the differentiated state leading to cancer. Bioessays. 1990 Apr;12(4):163–166. doi: 10.1002/bies.950120404. [DOI] [PubMed] [Google Scholar]
- Hori H., Takabayashi K., Orvis L., Carson D. A., Nobori T. Gene cloning and characterization of Pseudomonas putida L-methionine-alpha-deamino-gamma-mercaptomethane-lyase. Cancer Res. 1996 May 1;56(9):2116–2122. [PubMed] [Google Scholar]
- Hoshiya Y., Guo H., Kubota T., Inada T., Asanuma F., Yamada Y., Koh J., Kitajima M., Hoffman R. M. Human tumors are methionine dependent in vivo. Anticancer Res. 1995 May-Jun;15(3):717–718. [PubMed] [Google Scholar]
- Johnson B. A., Aswad D. W. Kinetic properties of bovine brain protein L-isoaspartyl methyltransferase determined using a synthetic isoaspartyl peptide substrate. Neurochem Res. 1993 Jan;18(1):87–94. doi: 10.1007/BF00966926. [DOI] [PubMed] [Google Scholar]
- Judde J. G., Ellis M., Frost P. Biochemical analysis of the role of transmethylation in the methionine dependence of tumor cells. Cancer Res. 1989 Sep 1;49(17):4859–4865. [PubMed] [Google Scholar]
- Kroes R. A., Erickson L. C. The role of mRNA stability and transcription in O6-methylguanine DNA methyltransferase (MGMT) expression in Mer+ human tumor cells. Carcinogenesis. 1995 Sep;16(9):2255–2257. doi: 10.1093/carcin/16.9.2255. [DOI] [PubMed] [Google Scholar]
- Kubota R., Kubota K., Yamada S., Tada M., Takahashi T., Iwata R., Tamahashi N. Methionine uptake by tumor tissue: a microautoradiographic comparison with FDG. J Nucl Med. 1995 Mar;36(3):484–492. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lishko V. K., Lishko O. V., Hoffman R. M. Depletion of serum methionine by methioninase in mice. Anticancer Res. 1993 Sep-Oct;13(5A):1465–1468. [PubMed] [Google Scholar]
- Liteplo R. G., Hipwell S. E., Rosenblatt D. S., Sillaots S., Lue-Shing H. Changes in cobalamin metabolism are associated with the altered methionine auxotrophy of highly growth autonomous human melanoma cells. J Cell Physiol. 1991 Nov;149(2):332–338. doi: 10.1002/jcp.1041490222. [DOI] [PubMed] [Google Scholar]
- Liteplo R. G. Reversion to a homocysteine-responsive phenotype in a human melanoma cell line is associated with diminished growth potential and increased methionine biosynthesis. Exp Cell Res. 1990 Feb;186(2):340–345. doi: 10.1016/0014-4827(90)90314-z. [DOI] [PubMed] [Google Scholar]
- Marathi U. K., Kroes R. A., Dolan M. E., Erickson L. C. Prolonged depletion of O6-methylguanine DNA methyltransferase activity following exposure to O6-benzylguanine with or without streptozotocin enhances 1,3-bis(2-chloroethyl)-1-nitrosourea sensitivity in vitro. Cancer Res. 1993 Sep 15;53(18):4281–4286. [PubMed] [Google Scholar]
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987 Jul 25;262(21):10035–10038. [PubMed] [Google Scholar]
- Mineura K., Sasajima T., Kuwahara N., Kowada M., Murakami M., Uemura K. (14C-methyl)-L-methionine uptake in rat brain tumors before and after treatment with the protein synthesis inhibitor cycloheximide. J Neurooncol. 1993 Mar;15(3):229–233. doi: 10.1007/BF01050068. [DOI] [PubMed] [Google Scholar]
- Pegg A. E. S-adenosylmethionine decarboxylase: a brief review. Cell Biochem Funct. 1984 Jan;2(1):11–15. doi: 10.1002/cbf.290020105. [DOI] [PubMed] [Google Scholar]
- Pieper R. O., Costello J. F., Kroes R. A., Futscher B. W., Marathi U., Erickson L. C. Direct correlation between methylation status and expression of the human O-6-methylguanine DNA methyltransferase gene. Cancer Commun. 1991 Aug;3(8):241–253. doi: 10.3727/095535491820873092. [DOI] [PubMed] [Google Scholar]
- Qian X., von Wronski M. A., Brent T. P. Localization of methylation sites in the human O6-methylguanine-DNA methyltransferase promoter: correlation with gene suppression. Carcinogenesis. 1995 Jun;16(6):1385–1390. doi: 10.1093/carcin/16.6.1385. [DOI] [PubMed] [Google Scholar]
- Schold S. C., Jr, Brent T. P., von Hofe E., Friedman H. S., Mitra S., Bigner D. D., Swenberg J. A., Kleihues P. O6-alkylguanine-DNA alkyltransferase and sensitivity to procarbazine in human brain-tumor xenografts. J Neurosurg. 1989 Apr;70(4):573–577. doi: 10.3171/jns.1989.70.4.0573. [DOI] [PubMed] [Google Scholar]
- Schold S. C., Jr, Kokkinakis D. M., Rudy J. L., Moschel R. C., Pegg A. E. Treatment of human brain tumor xenografts with O6-benzyl-2'-deoxyguanosine and BCNU. Cancer Res. 1996 May 1;56(9):2076–2081. [PubMed] [Google Scholar]
- Stern P. H., Hoffman R. M. Elevated overall rates of transmethylation in cell lines from diverse human tumors. In Vitro. 1984 Aug;20(8):663–670. doi: 10.1007/BF02619617. [DOI] [PubMed] [Google Scholar]
- Stern P. H., Hoffman R. M. Enhanced in vitro selective toxicity of chemotherapeutic agents for human cancer cells based on a metabolic defect. J Natl Cancer Inst. 1986 Apr;76(4):629–639. doi: 10.1093/jnci/76.4.629. [DOI] [PubMed] [Google Scholar]
- Sun Y., Kim H., Parker M., Stetler-Stevenson W. G., Colburn N. H. Lack of suppression of tumor cell phenotype by overexpression of TIMP-3 in mouse JB6 tumor cells identification of a transfectant with increased tumorigenicity and invasiveness. Anticancer Res. 1996 Jan-Feb;16(1):1–7. [PubMed] [Google Scholar]
- Tano K., Shiota S., Collier J., Foote R. S., Mitra S. Isolation and structural characterization of a cDNA clone encoding the human DNA repair protein for O6-alkylguanine. Proc Natl Acad Sci U S A. 1990 Jan;87(2):686–690. doi: 10.1073/pnas.87.2.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautt J. W., Anuszewska E. L., Koziorowska J. H. Methionine regulation of N-5-methyltetrahydrofolate: homocysteine methyltransferase and its influence on the growth and protein synthesis in normal, neoplastic, and transformed cells in culture. J Natl Cancer Inst. 1982 Jul;69(1):9–14. [PubMed] [Google Scholar]
- Tsujimura T., Zhang Y. P., Fujio C., Chang H. R., Watatani M., Ishizaki K., Kitamura H., Ikenaga M. O6-methylguanine methyltransferase activity and sensitivity of Japanese tumor cell strains to 1-(4-amino-2-methyl-5-pyrimidinyl)methyl-3-(2-chloroethyl)- 3-nitrosourea hydrochloride. Jpn J Cancer Res. 1987 Nov;78(11):1207–1215. [PubMed] [Google Scholar]
- Vanhamme L., Szpirer C. Spontaneous and 5-azacytidine-induced revertants of methionine-dependent tumor-derived and H-ras-1-transformed cells. Exp Cell Res. 1989 Mar;181(1):159–168. doi: 10.1016/0014-4827(89)90190-0. [DOI] [PubMed] [Google Scholar]
- Wang Y., Kato T., Ayaki H., Ishizaki K., Tano K., Mitra S., Ikenaga M. Correlation between DNA methylation and expression of O6-methylguanine-DNA methyltransferase gene in cultured human tumor cells. Mutat Res. 1992 Mar;273(2):221–230. doi: 10.1016/0921-8777(92)90083-f. [DOI] [PubMed] [Google Scholar]
- von Wronski M. A., Brent T. P. Effect of 5-azacytidine on expression of the human DNA repair enzyme O6-methylguanine-DNA methyltransferase. Carcinogenesis. 1994 Apr;15(4):577–582. doi: 10.1093/carcin/15.4.577. [DOI] [PubMed] [Google Scholar]
- von Wronski M. A., Harris L. C., Tano K., Mitra S., Bigner D. D., Brent T. P. Cytosine methylation and suppression of O6-methylguanine-DNA methyltransferase expression in human rhabdomyosarcoma cell lines and xenografts. Oncol Res. 1992;4(4-5):167–174. [PubMed] [Google Scholar]