Abstract
Analysis of the P. murina MSG gene family and expression-site locus showed that, as in P. carinii, P. murina MSG genes are arranged in head-to-tail tandem arrays located on multiple chromosomes, and that a variety of MSG genes can reside at the unique P. murina expression site. Located between the P. murina expression site and attached MSG gene is a block of 132 basepairs that is also present at the beginning of MSG genes that are not at the expression site. The center of this sequence block resembles the 28 basepair CRJE of P. carinii, but the block of conserved sequence in P. murina is nearly five times longer than in P. carinii, and much shorter than in Pneumocystis wakefieldiae. These data indicate that the P. murina expression-site locus has a distinct structure.
Keywords: Pneumocystis, mouse, antigen, variation, gene family, gene expression
Introduction
The fungal genus Pneumocystis contains multiple species including the causative agent of human Pneumocystis pneumonia, which afflicts individuals with impaired immune system function, such as Acquired Immunodeficiency Syndrome (AIDS) patients (Thomas, C. F., Jr. and Limper, A. H. 2004). Studying Pneumocystis organisms is difficult because they do not proliferate well in culture (Walzer, P. D. et al. 2001). Therefore, animal models have been the main source of organisms (Armstrong, M. Y. and Cushion, M. T. 1994; Dei-Cas, E. et al. 1998; Larsen, H. H. et al. 2002).
Pneumocystis murina, the species of Pneumocystis found in laboratory mice (Keely, S. P. et al. 2004), is of interest because the laboratory mouse provides an advanced animal model of host response to Pneumocystis infection (Wright, T. W. et al. 2001; Lund, F. E. et al. 2003; An, C. L., Gigliotti, F., and Harmsen, A. G. 2003; Qureshi, M. H., Harmsen, A. G., and Garvy, B. A. 2003; McAllister, F. et al. 2004; Empey, K. M. et al. 2004; Linke, M. et al. 2005; Qureshi, M. H., Empey, K. M., and Garvy, B. A. 2005; Linke, M. et al. 2006a; Linke, M. et al. 2006b). P. murina is a close relative of P. carinii and P. wakefieldiae, both found in rats, and a more distant relative of the human pathogen, P. jirovecii (Frenkel, J. K. 1976; Frenkel, J. K. 1999; Stringer, J. R., Cushion, M. T., and Wakefield, A. E. 2001; Stringer, J. R. et al. 2002; Keely, S. P. and Stringer, J. R. 2005; Redhead, S. A. et al. 2006). All four of these species feature an abundant surface protein called Major Surface Glycoprotein (MSG) (see Table 1), which has also been observed in Pneumocystis from ferrets, where it is known as gpA (Linke, M. J., Cushion, M. T., and Walzer, P. D. 1989; Tanabe, K. et al. 1989; Haidaris, C. G. et al. 1991; Nakamura, Y. et al. 1991; Haidaris, P. J. et al. 1992; Gigliotti, F. 1992; Stringer, S. L. et al. 1993; Kovacs, J. A. et al. 1993; Garbe, T. R. and Stringer, J. R. 1994; Wright, T. W. et al. 1994; Kitada, K., Wada, M., and Nakamura, Y. 1994; Wright, T. W. et al. 1995; Haidaris, C. G. et al. 1998).
Table 1.
Abbreviations referring to P. murina MSG gene family and its expression site
| Abbreviation | Meaning | Defining Characteristics |
|---|---|---|
| MSG | Major Surface Glycoprotein | The predominant protein found on the surface of Pneumocystis organisms. MSGs are approximately 120 kDa, are glycosylated and cysteine rich. There are many different isoforms of MSG, each encoded by a member of the MSG gene family. |
| UCS | Upstream Conserved Sequence | The conserved sequence found at the 5-prime end of every messenger RNA molecule encoding an MSG protein. |
| CRJE | Conserved Recombination Element | The conserved sequence found at the beginning of every open reading frame encoding an MSG protein. |
| ES | Expression Site | The locus to which an MSG gene must be attached in order to be transcribed. The ES includes the UCS locus and a presumptive promoter. |
Studies on other Pneumocystis species, primarily P. carinii, suggest that P. murina may use a family of MSG genes to produce antigenic variation in populations of the fungus dwelling in mice. P. carinii MSG is encoded by a multigene family, members of which are arranged as head-to-tail repeats located near the telomeres of all 17 chromosomes (Wada, M. et al. 1993; Kovacs, J. A. et al. 1993; Kitada, K., Wada, M., and Nakamura, Y. 1994; Wada, M. et al. 1995; Edman, J. C. et al. 1996; Stringer, J. R. and Keely, S. P. 2001; Cornillot, E. et al. 2002; Stringer, J. R. 2003; Stringer, J. R. 2005; Keely, S. P. et al. 2005). All indications are that only one MSG gene is expressed per P. carinii organism. Control of P. carinii MSG expression involves a single-copy locus called the expression site, or ES (Table 1). The majority of organisms in P. carinii populations appear to be haploid (Wyder, M. A., Rasch, E. M., and Kaneshiro, E. S. 1998). Because only one MSG gene can occupy the expression site at a time, the expression site system restricts transcription of the family to one gene per organism (Wada, M. et al. 1995; Edman, J. C. et al. 1996; Sunkin, S. M. and Stringer, J. R. 1996; Sunkin, S. M. and Stringer, J. R. 1997; Keely, S. P., Cushion, M. T., and Stringer, J. R. 2003). A large number of different MSG genes have been observed at the P. carinii expression site, suggesting that MSG genes can be moved to this locus by DNA recombination (Sunkin, S. M. and Stringer, J. R. 1997; Keely, S. P., Cushion, M. T., and Stringer, J. R. 2003). The mechanism of recombination is not known, but the possibility of a site-specific recombinase has been raised by the presence of a 28 basepair sequence called the Conserved Recombination Junction Element (CRJE) (Table 1). There is a copy of the CRJE at the junction between the expression site and adjacent MSG coding region. In addition to this copy, every MSG gene not at the expression site begins with a copy of the CRJE. Thus it is possible that recombination between the expression site and MSG genes at other loci is mediated by interactions between two copies of the CRJE. Whatever the mechanism, switching the MSG gene at the expression site discontinues expression of the previous resident MSG gene and activates expression of the new expression-site-linked MSG gene. Studies with antibodies that bind to few isoforms of MSG have shown that the protein encoded by the MSG gene at the expression-site locus is present in or on P. carinii cells (Schaffzin, J. K. and Stringer, J. R. 2004). Other studies using antibodies detected antigen variation between clusters of P. carinii in a rat lung (Angus, C. W. et al. 1996). Hence it would appear that P. carinii populations in the lung develop antigenic variation by switching the MSG gene that is at the expression site.
The P. carinii expression site encodes most of the 410 basepair Upstream Conserved Sequence (UCS) (Table 1), so named because it was found at the five prime ends of mRNAs encoding diverse MSGs (Wada, M. et al. 1995; Edman, J. C. et al. 1996; Sunkin, S. M. and Stringer, J. R. 1996; Sunkin, S. M. and Stringer, J. R. 1997; Sunkin, S. M. et al. 1998; Stringer, J. R. and Keely, S. P. 2001; Kutty, G., Ma, L., and Kovacs, J. A. 2001; Keely, S. P., Cushion, M. T., and Stringer, J. R. 2003; Stringer, J. R. 2003; Schaffzin, J. K. and Stringer, J. R. 2004; Stringer, J. R. 2005). The last 28 basepairs of the UCS are encoded by the CRJE. The UCS serves as the translation initiation site for production of an MSG precursor peptide (Sunkin, S. M. et al. 1998). The peptide encoded by the UCS appears to serve to send a precursor of mature MSG into the secretory pathway for transport to the cell surface (Sunkin, S. M. et al. 1998). The MSG on the surface lacks the amino acids encoded by the UCS, suggesting that these are removed by proteolysis in the endoplasmic reticulum (Sunkin, S. M. et al. 1998).
Given the similarities between species in the genus Pneumocystis, P. murina would be expected to contain a family of MSG genes that are expressed via an expression-site mechanism. However, this species might also be expected to have its own properties in this regard. Therefore, structural analysis of the expression-site locus of P. murina was necessary. Previously, a sequence that is approximately 62% identical to the P. carinii UCS was described at the 5’ end of a P. murina mRNA encoding an MSG (Haidaris, C. G. et al. 1998). Part of this sequence was shown to map to one chromosome of P. murina, suggesting that there is a single expression-site locus in this species (Haidaris, C. G. et al. 1998). The data presented herein characterize the locus encoding the P. murina UCS and provide more information about the MSG gene family in this species. These data show that the UCS locus of P. murina is present once per haploid genome and that a population of P. murina organisms can contain different MSG genes attached to the UCS locus. Between the genomic sequence encoding the UCS and the attached MSG gene, there is a sequence resembling the 28 bp CRJE of P. carinii, but sequence conservation extends substantially beyond the 28 bp CRJE-like sequence to encompass a block nearly five times longer than the CRJE of P. carinii. Multiple MSG P. murina genes were detected by hybridization, PCR and cloning. All but one chromosome contained MSG-related sequences. Head-to-tail tandem arrays of P. murina MSG genes were detected by PCR but no evidence of other tandem arrangements emerged. Sequence analysis detected 26 different MSG genes, which is only about one third as many as the number of MSG genes in P. carinii. Quantitative real-time PCR supported the hypothesis that the P. murina MSG gene family is smaller than the P. carinii MSG gene family.
Materials and Methods
Animal housing conditions
Mice were housed in microisolator cages in rooms with high efficiency particulate resistance filtered laminar airflow. Food and bedding material were autoclaved. All animal studies were performed in accordance with guidelines provided by the National Institutes of Health, University of Cincinnati, and the Veterans Affairs Medical Center.
Immunosuppression and infection
C3H/HeN Severe Combined Immunodeficiency (SCID) mice were immunosuppressed by the addition of dexamethasone (0.4 mg/ml) to their drinking water. Ampicillin (0.5 mg/ml) was also present in the drinking water to prevent bacterial infections. Immunosuppressed mice were inoculated with P. murina that had been previously isolated from C3H/HeN-SCID mice. To perform the inoculation, the animal was lightly anesthetized with halothane, a small feeding tube was inserted through the oral cavity into the trachea, and a volume of 0.05 ml of phosphate-buffered saline (PBS: 10 mM sodium phosphate, 138 mM NaCl, 2.7 mM KCl, pH 7.4) containing 107 P. murina was injected followed by 0.2 ml of air. After inoculation, mice were given water containing dexamethasone (0.4 mg/ml) and Ampicillin (0.5 mg/ml). Eight to 12 weeks post inoculation, mice were sacrificed and P. murina extracted. Control, non-inoculated immunosuppressed mice were included in each experiment. Control mice remained uninfected.
Isolation and cryopreservation of P. murina organisms
Lungs were removed, placed in PBS, minced and then homogenized using a Stomacher 80 (Tekmar, Inc., Gehanna, Ohio). The homogenate was filtered through gauze, washed in PBS, and P. murina were recovered by centrifugation at 3400 rpm. Red blood cells present in the pellet were lysed by incubation with 0.85 M NH4Cl in a 37°C water bath for 10 minutes. P. murina cells were collected by centrifugation at 3400 rpm for 10 minutes, washed in PBS, and collected by centrifugation. The supernatant was removed and the cells were suspended in 2 ml of PBS. To determine the number of P. murina in a preparation, three drops (10 µl per drop) of the cell suspension were placed on a glass slide and slides were allowed to dry in ambient air. The dry slides were stained with Diff Quik (Baxter Scientific, McGraw Park, Ill.). Ten random fields in each drop were examined under oil at 1000X magnification and P. murina nuclei were counted. Cells in the remaining preparation were collected by centrifugation in an microcentrifuge (Eppendorf model 5418) (Eppendorf North America) at 3400 rpm for 10 minutes at room temperature. Cells were suspended in 5 ml freezing media (RPMI media, 7.5% DMSO and 10% fetal calf serum) and frozen in liquid nitrogen. Cryopreserved organisms were stored in liquid nitrogen.
Preparation of P. murina DNA
Approximately 9 million cryopreserved organisms were placed in 0.2 ml of a solution containing 0.1% SDS, 0.1 M NaCl, 0.0025 M EDTA, 0.05 M Tris, pH 8.5, and 0.5 mg/ml proteinase K. The organisms were incubated for 2 hours at 55°C and then heated at 99°C for 10 minutes. The preparation was subjected to centrifugation at 10,000 ✕ g for 5 minutes at 20°C. The supernatant was transferred to a new tube and an equal volume of isopropanol was added. The sample was frozen at −70°C and DNA was collected by centrifugation at 10,000 ✕ g for 5 minutes at 20°C. DNA was dissolved in 25 microliters of water.
Isolation of P. murina RNA and production of cDNA
Infected lungs were flash frozen in liquid nitrogen and ground into a fine powder and stored at −70°C (Linke, M. et al. 2005). Approximately 50 mgs of frozen powdered lung tissue were placed in 1.0 ml Trizol® Reagent (Invitrogen, Carlsbad, CA) and total RNA was isolated according to the manufacturer’s directions. The RNA was treated with 1 unit of RNAase-free DNAase (Promega USA, Madison WI) for 30 minutes in the buffer supplied with the enzyme. The preparation was then subjected to phenol:chloroform extraction and RNA recovered by ethanol precipitation (Sambrook, J. F., Fristch, E. F., and Maniatis, T. 1989). The RNA concentration was estimated by absorbance of 260 nanometer light. cDNA was made from approximately 1 ug of RNA using the SuperScript™ II RNAase H Reverse Transcriptase (Invitrogen, Carlsbad, CA) according to the manufacturer’s directions.
Primer design
Primers are listed in Table 2 and their locations shown in Figure 1. All but primers p7, p8, p9 and p10, were designed to match a site in the UCS region of the published P. murina MSG cDNA sequence (accession number AF043102). Primers p7–p10 were designed to match sites conserved among members of the P. murina MSG gene family. To identify possible conserved sites, MSG genes from three other Pneumocystis species (P. carinii, P. wakefieldiae, P. jirovecii) were aligned to the published P. murina MSG cDNA sequence. Sites that were least variable across species were identified (data not shown) and primers matching the P. murina sequence corresponding to each of four of these sites (see p7, p8, p9 and p10 in Figure 1 and Table 2) were produced.
Table 2.
Sequences and positions of PCR primers.
| Name | Sequence | Position1 | Region2 |
|---|---|---|---|
| p1 | 5’:TTTCTTGATATCCGTCTCTTTTC:3’ | 1–13 | E1 |
| p2 | 5’:ATGAGGATTGCATTTTTTGCGC:3’ | 40–61 | E1 |
| p2.1 | 5’: GCAACTTAGTTATGTCTTGGGG :3’ | 69–90 | E1 |
| p2.2 | 5’:TATGAAAATTTTGGGTTGGATCC:3’ | 386–408 | E2 |
| p2.2A | 5’: GGATCCAACCCAAAATTTTCATA :3’ | 408–386 | E2 |
| p3 | 5’:ATTATGATGGTTTCTTTTCTGAAG:3’ | 575–608 | E2 |
| p3A | 5’:CTTCAGAAAAGAAACCATCATAAT:3’ | 608–575 | E2 |
| P3.1 | 5’:TTCTCCTCTGAAGATTATTCTCA :3’ | 577–599 | E2 |
| p4 | 5’:AGAGGAAGTCGCCCAGAAAG:3’ | 649–668 | E2 |
| p5 | 5’:CGCCGCCTCTTTCTGGGCGA:3’ | 676–657 | E2 |
| p6 | 5’:ATGGCACAGCCGGTTAAGAG:3’ | 657–675 | E2 |
| p6.1 | 5’:AYCTCATCATTTCCTGCTGCT:3’ | 712–692 | E2 |
| p7 | 5’:TAATTCTTTGCATTTTTCTCCTTC:3’ | 896–873 | E2 |
| p8 | 5’:AATAGACATTTTTCTTCATATTTTCT:3’ | 1003–978 | E2 |
| p9 | 5’:GGAGTGATTGTTATGGGAATGTT:3’ | 4038–4060 | E2 |
| p10 | 5’:AATGAGGTGTAGATTTTTAATCTAG:3’ | 4143–4167 | 3’-UTR |
Reference sequence was Genbank sequence AF043102
Location of primer binding site: E1, exon 1; E2, exon2; MSG, major surface glycoprotein codon region; UTR, MSG gene 3-prime untranslated region.
Figure 1. Map of the P. murina expression-site locus.
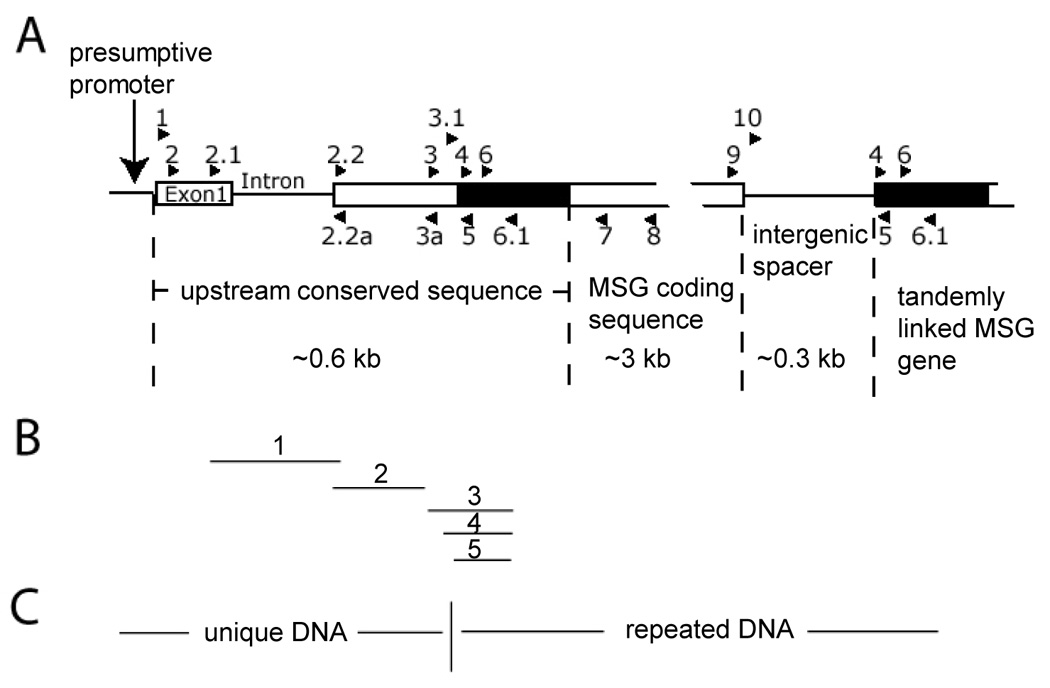
A) The map was constructed primarily from information obtained in the studies presented herein. The map begins upstream of the sequence analyzed. Presumably, there is a promoter of transcription located in this region. Exon 1 marks the beginning of the sequence that encodes the Upstream Conserved Sequence (UCS), which is the sequence found at the beginning of messenger RNAs encoding diverse MSGs. DNA encoding the UCS extends from exon 1, which is unique in the genome, through the intron into a second block of unique coding DNA. This block of unique DNA (exon 1-intron-beginning of exon 2) constitutes the expression site. Adjacent to the expression site is a block of non-unique DNA called the CRJE (see Table 1), which is represented by a black rectangle. The 3-prime end of the UCS coincides with the 3-prime end of the CRJE. Other copies of the CRJE are located at the beginning of MSG genes not at the expression site. Hence the beginning of the UCS is encoded by unique DNA, and the end is encoded repeated DNA. The sequence encoding the UCS is followed by a sequence encoding an MSG protein. Two tandem MSG-encoding sequences are shown. The first is directly attached to the expression site; the second lies downstream of the first, and is separated from the UCS-linked MSG gene by approximately 300 bp (intergenic spacer). It should be noted that while tandem gene pairs are demonstrated by the data presented herein, it is not yet known if a tandem gene pair exists at the expression site. Locations and names of PCR primers are indicated by numbered arrowheads. See Table 2 for primer sequences and locations. B) The five numbered lines mark regions studied by real-time PCR. C) The region to the left of the line contains a DNA sequence that is present only once in the haploid genome of P. murina. The region to the right of the line contains a DNA sequence that is present in multiple copies in the genome.
PCR amplification
When performed in order to produce cloned amplicons, PCR was performed under the following conditions: 95°C for 3 minutes, then 40 cycles of incubation at 95°C for 10 seconds, 45°C for 30 seconds, and 72°C for 30 seconds, then 72°C for 5 minutes, storage at 4°C. Reaction volumes were 25 microliters containing 100 mM each of dATP, dCTP, dGTP, dTTP, 1 U of Tfl polymerase (Epicenter, Madison, WI), 1.5 mM MgCl2, and 20 ng each of primer. Approximately 3 nanograms of P. murina genomic DNA was added to each reaction.
Real-time PCR was performed with a Cepheid thermocycler (Sunnyvale, CA) under the following conditions: 95°C for 15 seconds, annealing temperature for 15 seconds, and 72°C for 15 seconds. The optics were set to detect SYBR Green fluorescence. The concentrations of deoxyribonucleotide triphosphates were the same as those described above. The reactions contained Taq polymerase (1 unit per 25µl) (Promega USA, Madison WI), 5 mM MgCl2, 1:1 of a 1:10,000 dilution of SYBR Green (BioWhitaker Molecular Applications) and 20 ng of each primer. Approximately 3 nanograms of P. murina genomic DNA was added to each reaction. The annealing temperatures used for primer pairs were as follows: 50°C for p2.1/p2a, p2/p3a and p3.1/p6.1, 55°C for p3/p6.1 and p4/p6.1. The performance of each primer pair was assessed by linear regression analysis of amplification kinetics of reactions containing known amounts of plasmid carrying the amplification target. For each experiment, cycle threshold (CT) was plotted versus amount of plasmid DNA. For example, Figure 5A shows a scatter plot of data from multiple trials performed with two primer pairs, p3/p6.1 and p4/p6.1. The figure shows that the CT values correlated (r² = 0.99) with the amount of plasmid over several logs of plasmid DNA concentration (Table 4). The amplification efficiency ranged from 71 to 82% (Table 4).
Figure 5. Quantitative real-time PCR of the MSG expression site.
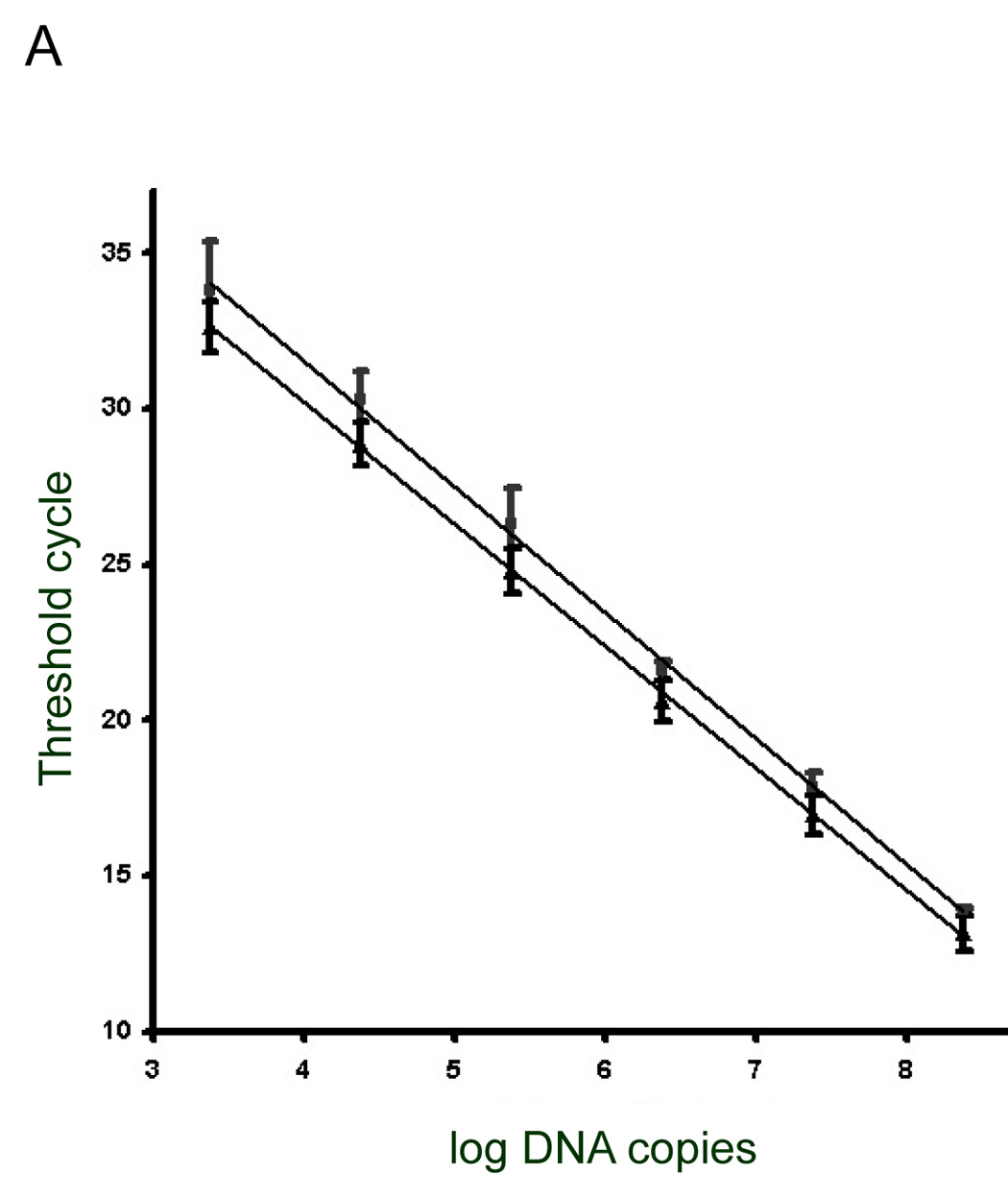
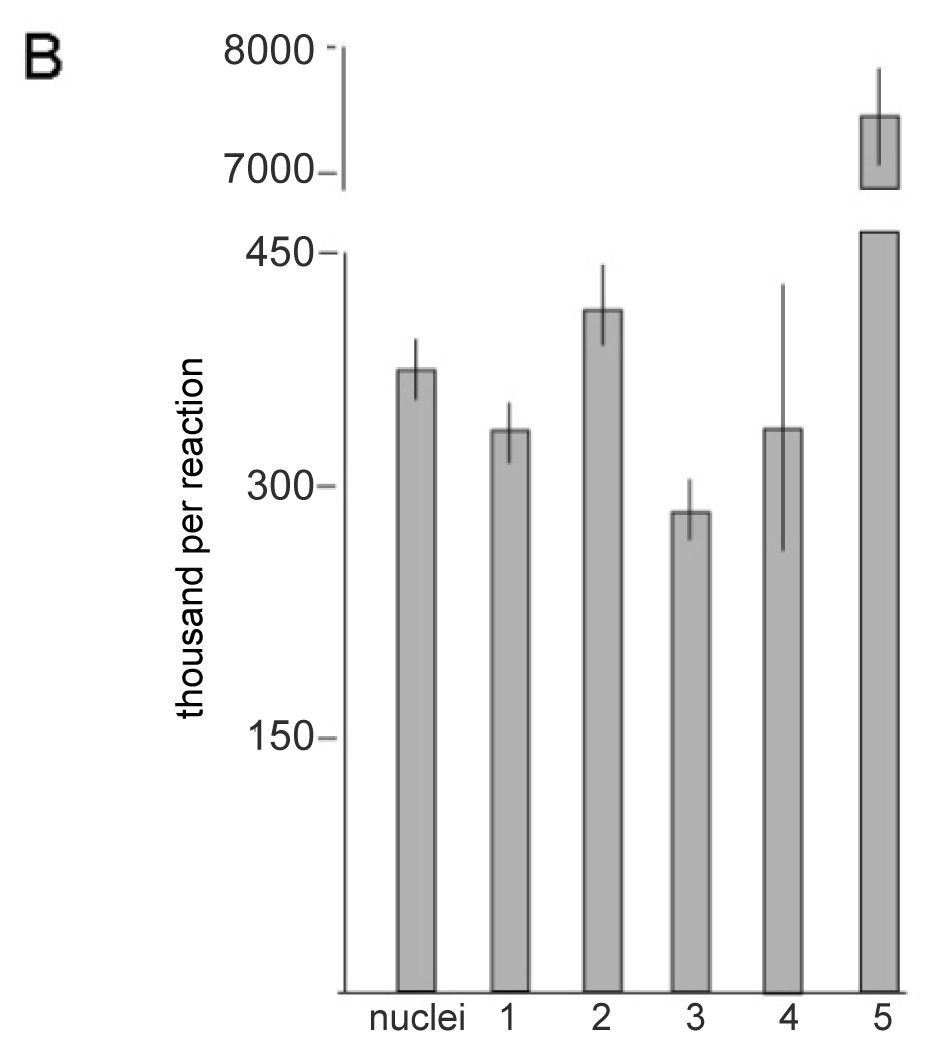
A) Examples of scatter plot data from plasmids used to establish the relationship between target copy-number (DNA copy number) and PCR cycles required to obtain detectable amplicons (threshold cycle number). Boxes are data points obtained using primers p3 and p6.1. Triangles are data points obtained using primers p4 and p6.1. B) Calculated copy-numbers of five UCS regions compared to the number of P. carinii nuclei present in each PCR. The bar labelled “nuclei” indicates the number of P. murina nuclei added to each reaction. Nuclei were counted twice and the line at the top of the bar indicates the standard deviation. Regions 1, 2, 3, 4 and 5 (see Figure 1B), were amplified with primers p2.1 plus p2.2a, p2.2 plus p3a, p3 plus p6.1, and p3.1 plus p6.1, and p4 plus p6.1, respectively. Three independent PCR reactions were performed with a given primer pair. Lines at the tops of bars indicate standard deviations.
Table 4.
Information about real-time PCR
| Regiona | Product length (bp) | Upper primera | Lower primera | Efficiency (%)b | Amplicon Melting temperature (°C) | r²c |
|---|---|---|---|---|---|---|
| 1 | 320 | p2.1 | p2.2A | 80.34 | 80 | 0.9941 |
| 2 | 223 | p2.2 | p3A | 75.39 | 85 | 0.9972 |
| 3 | 149 | p3 | p6.1 | 71.28 | 89 | 0.9968 |
| 4 | 124 | p3.1 | p6.1 | 77.99 | 89 | 0.9950 |
| 5 | 85 | p4 | p6.1 | 81.76 | 88 | 0.9975 |
See Figures 1A and 1B and Table 1
Efficiency (%) = (10−1/slope − 1) × 100
Residuals computed by linear regression analysis of data points produced from serial dilutions of template DNA.
Sequence analysis
Amplicons were cloned into the plasmid TOPO 4.0 (Invitrogen, Carlsbad, CA), which was introduced into the strain of E. coli provided with the vector. DNA sequences were determined by the Sequencing Facility at the University of Cincinnati College of Medicine. Most sequences were determined from both strands. Sequences were aligned using DNAMAN software (Lynnon BioSoft, Vaudreuil, Quebec, Canada) using the default settings. The alignments were optimized by introducing a limited number of gaps. The relatedness of sequences was evaluated and depicted using Mega 3.1 software (Kumar, S., Tamura, K., and Nei, M. 2004). In assessing relatedness, all codon positions were used and assigned equal weight, however, gaps in pairwise sequence alignments were ignored. Distances between sequences (p distances) were calculated by dividing the number of mismatched positions by the total number of positions, with apparent transitions and transversions receiving equal weight. Computer analysis of UCS-encoded peptides was performed with artificial neural networks and hidden Markov models provided at the SignalP 3.0 website (http://www.cbs.dtu.dk/services/SignalP/). The sequence of the P. murina UCS locus is listed under accession number EF158849.
CHEF electrophoresis
P. murina organisms were passed through two 10µm pore size filters (Mitex; Millipore Corp., Bedford, Mass) and extracellular DNA was digested with DNAase I (Boehringer Mannheim Biochemicals, Indianapolis, IN) at 10 µg/ml in a solution of 150 mM NaCl-10 mM MgCl2-10 mM Tris at pH 7.2 for 30 min at 37°C. The DNAase was inhibited by removing magnesium ions by washing with 125–250 mM EDTA (Hong, S. T. et al. 1990; Cushion, M. T. et al. 1993; Cushion, M. T. et al. 2001). Organisms were embedded in 0.8% low-melt agarose (Boehringer-Mannheim), then lysed and deproteinized by incubation at 55°C for 24–48 hr in 0.45 M EDTA, 0.01 M Tris containing 1% N-lauroylsarcosine (Sigma Chemical Co., St. Louis, MO) and 0.25 mg proteinase K (Boehringer-Mannheim) per ml. Digested samples were stored at 4°C in 0.5 M EDTA. Gels for Contour Clamped Homogeneous Electrical Field (CHEF) electrophoresis contained 1% FMC SeaKem GTG-agarose (SeaKem, Rockland, ME) prepared in 0.5X TBE (45 mM Tris HCl, 45 mM boric acid, 1.25 mM EDTA) for a total volume of 200 ml and final dimensions of 14 by 21 cm. Electrophoresis was performed using either a Bio-Rad CHEF DR II or CHEF DR III apparatus. Gels were run for 104 to 144 hrs, at 14°C, in 0.5X TBE at 3.8V/cm with a 50 second initial pulse that was gradually increased to 100 seconds. DNA bands were transferred by capillary action to positively charged nylon blots under neutral conditions, and UV-crosslinked to the membranes, as described previously (Hong, S. T. et al. 1990; Cushion, M. T. et al. 1993; Cushion, M. T. et al. 2001).
Southern blot hybridization
To generate the UCS probe, approximately 3 nanograms of genomic DNA from P. murina were subjected to amplification with primers p1 and p5, as described above. The amplicon was cloned into TOPO 4.0 (Invitrogen, Carlsbad, CA) and sequenced. The plasmid was cut with restriction endonucleases Eco R1 and Bam H1 to release a 380 bp ‘EB’ fragment containing expression site exon 1, intron, and 20 bp beyond the end of the intron. The ‘EB’ fragment was gel-purified by electrophoresis in 2% NuSieve GTG agarose (BioWhittaker Molecular Applications, Rockland, ME) (Sambrook, J. F., Fristch, E. F., and Maniatis, T. 1989)and recovered from the gel using a QIAquick Gel Extraction kit (Qiagen, Inc., Valencia, CA). The fragment was made radioactive using a Prime-it II Random Primer Labeling Kit (Stratagene, La Jolla, CA). Radioactive DNA was denatured by boiling for 5 minutes and added to a vessel containing Rapid-Hybe (Amersham) and a Southern blot. Hybridization was performed at 50°C for 18 hours. After hybridization, membranes were washed three times at 50°C in 2X SSC (0.3M NaCl, 0.03M NaCitrate) and 0.1% sodium dodecyl sulfate (SDS). Bound radioactive probe was detected by autoradiography.
To generate the MSG probe, primers p6 and p7 were utilized to produce an amplicon from approximately 3 nanograms of genomic DNA from P. murina. PCR was performed under the following conditions: 95°C for 3 minutes, then 40 cycles of incubation at 95°C for 10 seconds, 45°C 30 seconds, and 72°C for 30 seconds, then 72°C for 5 minutes, storage at 4°C. Reaction volumes were 25 microliters containing 100 mM each of dATP, dCTP, dGTP, dTTP, 1 U of Tfl polymerase (Epicenter, Madison, WI), 1.5 mM MgCl2, and 20 ng each of primer. Amplification with primers p6 and p7 was expected to yield a 325 bp product, but a product closer to 1 kb in size was obtained. Sequencing showed that the amplicon ended at a point 716 downstream of the expected p7 priming site. Alignment of the 1 kb sequence to the P. murina MSG cDNA sequence listed under accession number AF043102 showed that the 1 kb amplicon was 94% identical to a P. murina MSG gene and identified a site where primer p7 matched 19 of24 bases. These data explained the source of the 1 kb amplicon and established that it was suitable for use as hybridization probe. The entire plasmid was made radioactive using a Prime-it II Random Primer Labeling Kit (Stratagene, La Jolla, CA), denatured with heat, and incubated with the CHEF blot in Rapid-Hybe (Amersham) at 50°C for 18 hours. After hybridization, membranes were washed three times at 50°C in 2X SSC (0.3M NaCl, 0.03M NaCitrate) and 0.1% sodium dodecyl sulfate (SDS). Bound radioactive probe was detected by autoradiography.
To generate the kex1 probe, approximately 3 nanograms of genomic DNA from P. murina were subjected to PCR amplification with primer KEX-upper (5’:GGGATAAATCTTGGAAGGAAA:3’) and primer KEX-lower (5’:GAACCCGAATATGTAGAAGCA: 3’) which bind nucleotide positions 979 to 999 and 1158 to 1178, respectively, of the sequence listed under accession no. AF093132. PCR was performed under the following conditions: 95°C for 3 minutes, then 40 cycles of incubation at 95°C for 10 seconds, 55°C 30 seconds, and 72°C for 30 seconds, then 72°C for 5 minutes, storage at 4°C. Reaction volumes were 25 microliters containing 100 mM each of dATP, dCTP, dGTP, dTTP, 1 U of Tfl polymerase (Epicenter, Madison, WI), 1.5 mM MgCl2, and 20 ng each of primer. The amplicon was cloned into TOPO 4.0 (Invitrogen, Carlsbad, CA) and the identity of the insert in the plasmid was confirmed by sequencing. A radioactively labeled DNA probe was prepared as described above. Hybridization was performed in a solution containing 6XSSC (0.9M NaCl, 0.09M sodium citrate) 5X Denhardt's reagent (water containing 1% (weight:volume) Ficoll (Type 400) (Pharmacia), Polyvinylpyrolidone (Sigma) and BSA (Fraction V) (Sigma)) 0.5% SDS, and 100 ug of sheared herring sperm DNA per ml at 65°C for 18 hours(Cushion, M. T. et al. 1993). Unbound probe was removed by 3 washes with 2XSSC, 0.1% SDS at 60°C. Bound probe was detected by autoradiography as described (Cushion, M. T. et al. 1993).
Results
Sequence of the locus encoding the P. murina UCS
A previous study reported that a P. murina transcript encoding an MSG began with a sequence that was similar to the UCS from P. carinii (Haidaris, C. G. et al. 1998). To determine the structure of the locus encoding this UCS-like RNA sequence, the locus was isolated by PCR using primers p1 and p5 (Table 2, Figure 1), which amplified a 676 bp segment of the P. murina genome (Figure 2).
Figure 2. Sequence at the P. murina expression-site locus.
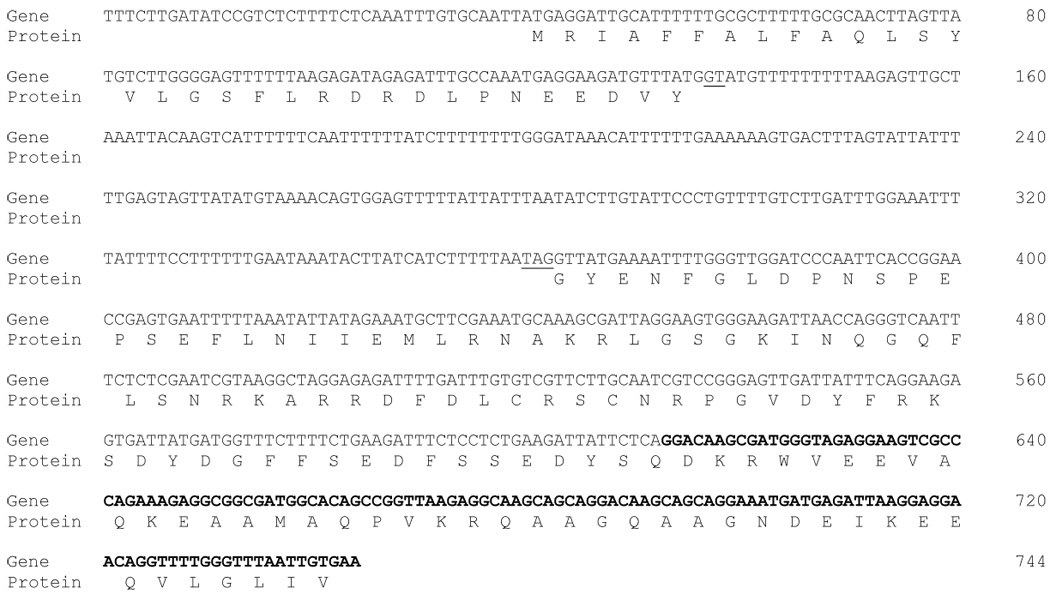
The DNA sequence shown encodes the conserved RNA sequence found at the beginning of messenger RNA molecules that encode different MSG proteins. The sequence begins in a region of exon 1 that is transcribed but presumably untranslated and ends at the end of the CRJE. The gap in the protein sequence marks the intron. Canonical splice acceptor and donor sequences are underlined. Nucleotides in bold print constitute the CRJE. Numbers at the right end of each line of nucleotide sequence indicate the cumulative number of nucleotides in the sequence to that point.
Comparison of amplified genomic DNA sequence to the sequence at the beginning of the previously described P. murina transcript identified a 227 bp segment present only in the genomic sequence. This segment began and ended with canonical splice donor and acceptor sites, GT and TAG, respectively (Figure 2), suggesting that the expression-site locus of P. murina contains an intron, as is the case in other species of Pneumocystis (Wada, M. et al. 1995; Sunkin, S. M. and Stringer, J. R. 1996; Schaffzin, J. K. and Stringer, J. R. 2000; Kutty, G., Ma, L., and Kovacs, J. A. 2001). Putative splice donor and acceptor sequences were similar to those of UCS introns from three other Pneumocystis species and to the consensus sequences for 19 different intron-containing genes (Table 3).
Table 3.
Comparison of splice donor and splice acceptor regions in four Pneumocystis species.
| Exon 1 | Intron Donor | Intron Acceptor | Exon 2 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| −3 | −2 | −1 | +1 | +2 | +3 | +4 | +5 | +6 | +6 | +5 | +4 | +3 | +2 | +1 | −1 | −2 | −3 | |
| D | W | D | G | T | W | W | D | W | W | W | W | Y | A | G | D | H | W | |
| PC | A | T | G | G | T | G | A | G | T | T | T | C | T | A | G | A | A | G |
| PW | A | T | G | G | T | A | T | G | T | C | T | A | C | A | G | A | T | T |
| PJ | G | A | G | G | T | A | T | G | T | T | G | G | C | A | G | A | T | C |
| PM | A | T | G | G | T | A | T | G | T | T | A | A | T | A | G | G | T | T |
Abbreviations are: A, adenine; C, cytosine; G, guanine; T, thymine; N, nucleotide; D, not cytosine; W, adenine or thymine; Y, thymine or cytosine; H, not guanine. The consensus sequence is from Thomas et al. (Thomas, C. F., Jr., Leof, E. B., and Limper, A. H. 1999)
Removal of the intron from the genomic UCS sequence generated an open reading frame encoding a 130 amino acid polypeptide 40–45% identical to the UCS-encoded polypeptides of P. carinii and P. wakefieldiae. Computer analysis of these three predicted peptides suggested that each of the three UCS-encoded peptides can function to send a nascent MSG polypeptide into the endoplasmic reticulum, consistent with results from previous experiments, where the P. carinii UCS-encoded peptide was shown to function as a signal peptide in insect cells (Sunkin, S. M. et al. 1998).
To confirm that the 227 bp segment functions as an intron, 8 additional UCS-MSG cDNA sequences were produced (data not shown). These data confirmed that splicing occurs. The splice donor site lies between bases 97 and 98 when base 1 is the A residue in the AUG codon that presumably initiates translation (Figure 2). This splice site is exactly the same distance from the AUG codon as it is in P. carinii, P. jirovecii and P. wakefieldiae.
The P. murina genomic UCS sequence matched that of P. carinii at 50% of positions. The coding regions were more similar (58% identity) than the introns (45% identity) (Figure 3). The 227 bp P. murina intron is longer than the P. carinii intron, which varies between 150 and 162 bp in length, but sequences present at the beginning and end of the P. murina intron were similar to those in the P. carinii intron (Figure 3). The P. murina intron is approximately the same length as the intron in the UCS locus of P. wakefieldiae (220 bp), and these two introns were approximately 50% identical (alignment not shown).
Figure 3. Comparison of expression site loci in P. murina and P. carinii.
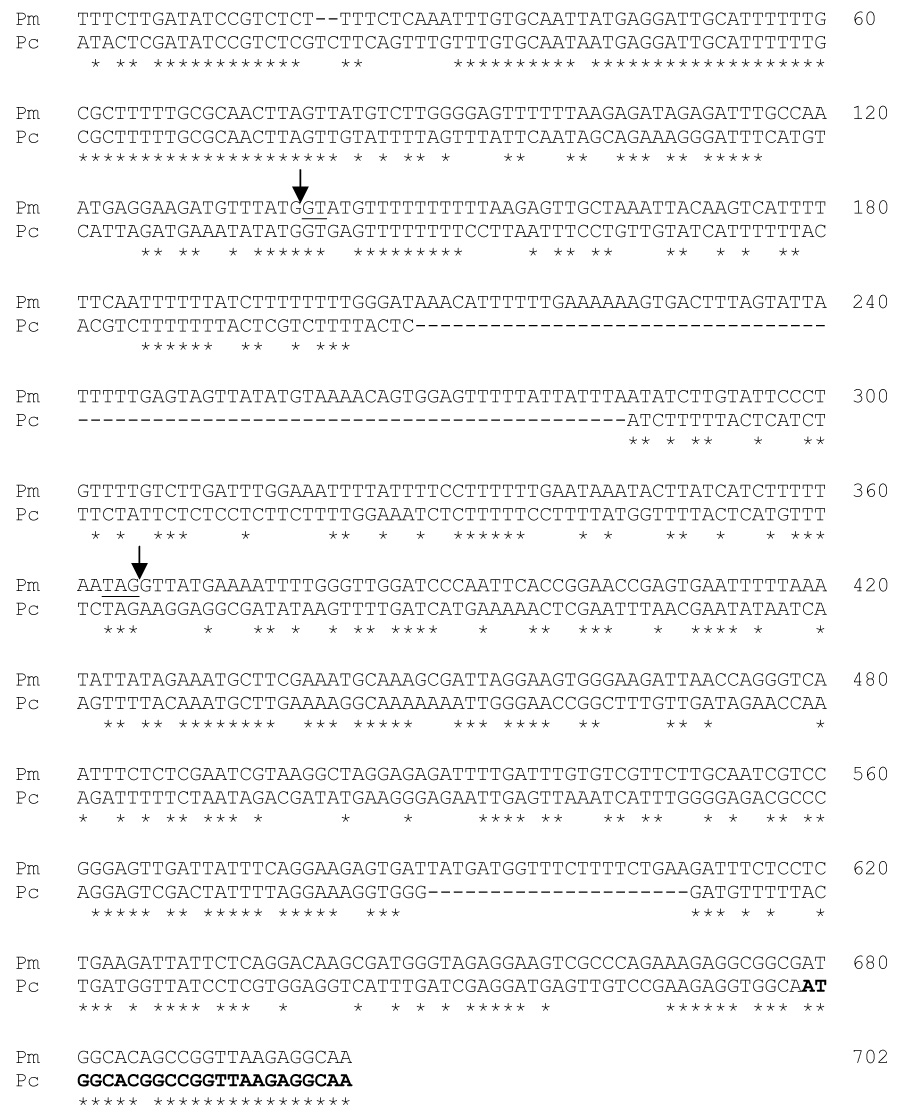
Pm and Pc indicate sequences from P. murina and P. carinii, respectively. The P. carinii sequence is from accession no. D82031. Canonical splice acceptor and donor sequences are underlined and splice points marked by arrows. Asterisks indicate positions where the two sequences match. The alignment ends at the end of the P. carinii CRJE, which is indicated by nucleotides in bold print. Numbers at the right indicate the cumulative number of sites in the alignment to that point.
Nearly 80% of the bases in the P. murina intron were either adenine (A) or thymidine (T), and the intron contained numerous A-rich simple repeats. For example, there were fifteen tracts comprised of consecutive adenine residues (ranging from 3 to 9 bases in length). Altogether, the intron contained twenty-three mononucleotide tracts, averaging 4.5 ± 1.7 (S.E.) nucleotides per tract. These tracts comprised forty-six percent of the intron. Mononucleotide tracts tend to be polymorphic within a species (Jonsson, A. B., Nyberg, G., and Normark, S. 1991). Therefore the intron of the P. murina UCS locus might have utility as a marker for population genetics studies.
UCS exon 1 and intron mapped to a single chromosome
Previously published data had shown that a 300 bp segment from the UCS part of a P. murina cDNA hybridized to a single P. murina chromosome, suggesting that the P. murina UCS is encoded by a single locus, as is the case in P. carinii (Lee, L. H. et al. 2000). To test this hypothesis and to determine if the intron within the UCS were single-copy in the genome, hybridization experiments were performed using exon 1 and the intron of the P. murina UCS as probe
Figure 4 (lane 4) shows that only one P. murina chromosome, approximately 440 Kb in size, hybridized strongly to the exon1-plus-intron probe, suggesting that the genome of P. murina contains a single copy of UCS exon 1 and intron. Later, quantitative PCR experiments, described below, confirmed the single-copy hypothesis.
Figure 4. Hybridization mapped the sequence encoding UCS to a single P. murina chromosome.
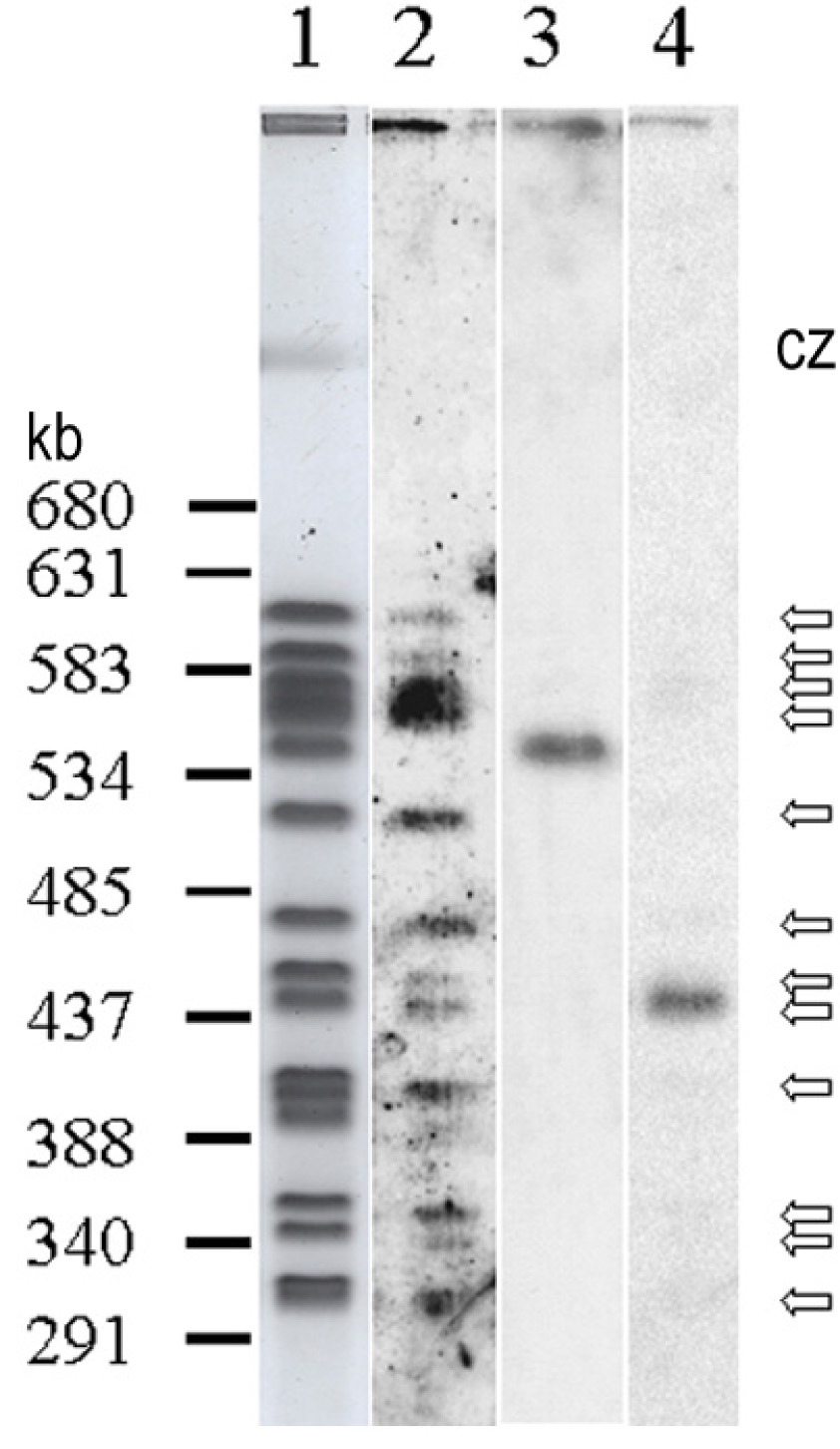
Lane 1: Negative image of Sybr-gold stained chromosomes resolved by CHEF. Lanes 2–4, autoradiograms showing chromosomes that hybridized to probes for MSG, kex1 and UCS, respectively. Numbers to the left of lane 1 indicate locations of markers ranging from 291 to 680 kilobasepairs (markers not shown). Open arrows to the right of lane 4 indicate chromosome bands scored as positive for the MSG probe. CZ indicates the compression zone, where DNA from the mouse host lung is located (visible in lane 1).
By contrast with the results obtained with the UCS exon1-intron probe, a probe that contained a 1 kb portion of a P. murina MSG gene hybridized to 12 chromosomes (Figure 4, lane 2). Some P. murina chromosomes produced stronger MSG hybridization signals than others, suggesting variation in either the number or sequences of the MSG gene family members. Hybridization conditions were nonstringent to allow probes to bind to MSG targets that were up to 20% different from the probe sequence. While conditions were not highly stringent, they were stringent enough to prevent indiscriminate probe binding. There was no signal from the 540 kb chromosome and none from the compression zone (compare lanes 1 and 2). In addition, the P. murina MSG probe did not hybridize to P. carinii chromosomes (data not shown).
The lack of MSG signal from the 540 kb chromosome was unexpected. To confirm that this result was not an artifact, the blot was hybridized to a P. murina kex1 probe because this probe had been reported to map to this chromosome (Lee, L. H. et al. 2000). The kex1 probe hybridized strongly to the 540 kb chromosome (lane 3), showing that the lack of signal when the MSG probe was applied was not due to an artifact of the experiment such as lack of transfer of this chromosome from the gel to the membrane.
Determining the copy number of the sequences at the UCS locus and mapping the boundary between unique and repeated DNA
The sequence downstream of UCS exon 1 was scanned for copy-number by quantitative real-time PCR using primer pairs that targeted five regions, 1, 2, 3, 4, and 5 (see Figure 1 and Table 2). Figure 5A shows the results of control experiments using plasmid DNA, which showed that target copy-number and rate of amplification were linearly related, as expected. Figure 5B shows the calculated copy-numbers of the five target regions compared to the number of P. carinii nuclei present in each PCR. These data showed that regions 1, 2, 3, and 4 were each present once per nucleus, consistent with there being a single-copy per genome. (Most P. carinii nuclei contain a haploid complement of DNA (Wyder, M. A., Rasch, E. M., and Kaneshiro, E. S. 1998)). The results obtained with regions 1–4 were similar to those obtained in control experiments targeting two other regions known to be single-copy in the P. murina genome (data not shown). By contrast with regions 1–4, region 5 amplified 19–23 times faster than would be expected for a single-copy target. Regions 3, 4 and 5 shared the same downstream primer. Therefore, rapid amplification of region 5 was due to repetition of the site for its upstream primer, p4. Thus, these data mapped the boundary between unique and repeated DNA to the 15 bp region between primers p3.1, which was the upstream primer site used to amplify region 4, and primer p4.
Diversity of MSG genes adjacent to the UCS locus
Populations of P. carinii tend to be diverse downstream of the UCS locus (Keely, S. P., Cushion, M. T., and Stringer, J. R. 2003). A PCR strategy was employed to examine sequence diversity at UCS-MSG junctions in P. murina. To increase the chance of amplifying UCS-MSG junctions that differ with respect to MSG, downstream primers p7 and p8 were designed to bind to sites expected to be present in multiple P. murina MSG genes (see Methods).
Six polymerase chain reactions were performed, each with one of three UCS-specific primers (p1, p2, or p3) paired with either p7 or p8 (Figure 1A). Twenty-eight cloned amplicons were sequenced and ten different MSG sequences were obtained. All of the MSG sequences contained an open reading frame continuous with the spliced version of the expression-site sequence. The average variation between MSG sequences was 8% (range: 1–13%).
Mapping the upstream border of the CRJE and detection of tandem MSG genes
In P. carinii, the CRJE is a 28 bp highly conserved sequence that is at the beginning of all MSG genes that are not at the expression site. A copy of the CRJE is also located between the expression site and an attached MSG gene (Wada, M. et al. 1995; Wada, M. and Nakamura, Y. 1996). While all Pneumocystis species examined so far have a sequence element homologous to the P. carinii CRJE, the size and sequence of the CRJE can vary among species (Schaffzin, J. K. and Stringer, J. R. 2000; Kutty, G., Ma, L., and Kovacs, J. A. 2001).
To characterize the upstream border of the P. murina CRJE, it was necessary to analyze the regions upstream of MSG genes that are not at the expression site. PCR presented itself as a possible tool for this purpose because MSG genes are arranged as head-to-tail tandem repeats in other species of Pneumocystis (Stringer, S. L. et al. 1991; Stringer, S. L. et al. 1993; Sunkin, S. M., Stringer, S. L., and Stringer, J. R. 1994; Garbe, T. R. and Stringer, J. R. 1994; Wada, M. and Nakamura, Y. 1994; Schaffzin, J. K., Garbe, T. R., and Stringer, J. R. 1999). This arrangement allows one to amplify the regions between MSG genes using an upstream primer that binds to a site conserved at the three prime ends of MSG genes, and a downstream primer that binds to a site conserved at the five prime ends of MSG genes.
Primer p9 (Table 2) binds to a sequence expected to be conserved at the 3-prime ends of P. murina MSG genes because this part of P. carinii MSG genes is highly conserved (Figure 1A). Primer p5 binds just upstream of the sequence that resembles the P. carinii CRJE (Figure 1A). This primer pair produced amplicons containing approximately 500 bp (not shown). Six cloned amplicons were sequenced, which showed that the procedure amplified at least 6 different tandem MSG gene pairs (data not shown). Similar data were obtained using a different upstream primer, primer p10, which binds in the 3’ untranslated regions of MSG genes (Figure 1A). In this case, five cloned amplicons were sequenced and five different sequences found (data not shown).
Comparison of the 11 amplified sequences to the sequence of the expression-site locus showed that the first 23 bp immediately upstream of the p5 primer site were identical to those at the expression site (Figure 6). Beyond these 23 bp, however, the sequences in the amplicons diverged from the expression site sequence. The divergence point is adjacent to the last nucleotide at the binding site for the primer p3.1 (Figure 6), which amplified the expression site with single-copy kinetics (Figure 5B). These data mapped the upstream border of the CRJE at the nucleotide level.
Figure 6. Comparison of expression-site to sequences (ES) upstream of MSG genes not at the expression site.
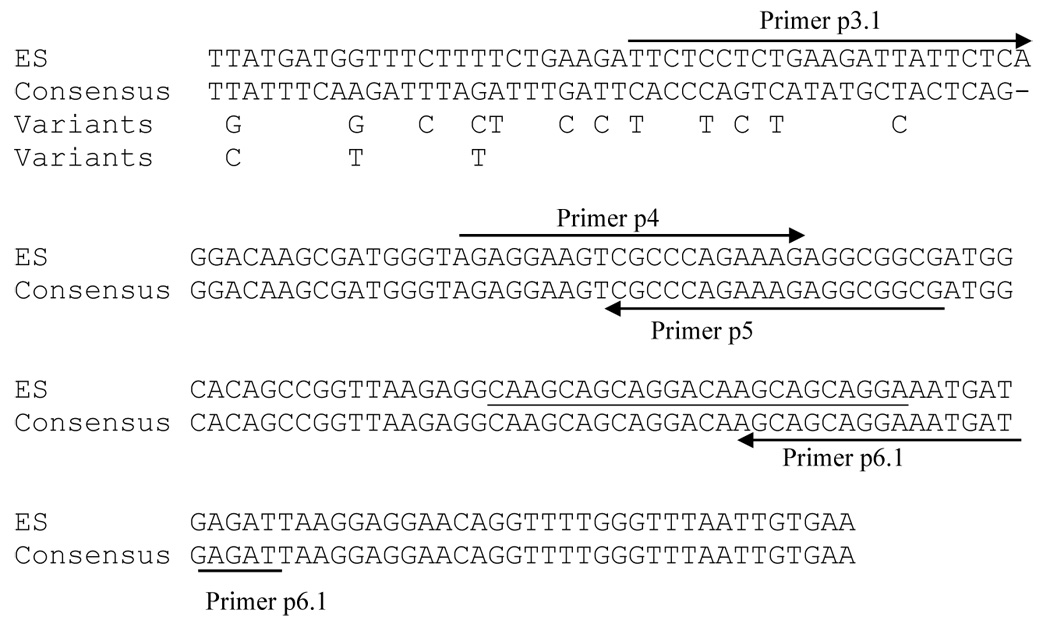
The top line of sequence is from the expression site. The second line is the consensus sequence derived from regions upstream of 11 MSG genes that are not at the expression site. The dash indicates a gap in the sequence alignment. The third and fourth lines show sequence polymorphisms in the regions upstream of MSG gene that are not at the expression site. The locations of the sequences that bind PCR primers p3.1, p4, p5 and p6.1 are shown by arrows. The 24 bases underlined contain 2 copies of a 12 base repeat. A third copy of this repeat was present in some sequences.
PCR using either primer p5 alone or primer p9 alone failed to produce amplicons, suggesting that head-to-head and tail-to-tail tandem MSG genes either do not exist in the P. murina genome, or are too far apart to be detected by PCR as performed.
Mapping the downstream border of the CRJE and assessment of MSG gene family complexity
Experiments with primer p8 (Figure 1A, Table 2) had indicated that this primer was capable of supporting amplification of multiple members of the P. murina MSG gene family, and would therefore be suited to the purpose of mapping the downstream border of the CRJE.
Genomic DNA between primers p4 and p8 was amplified and 56 cloned amplicons were sequenced. These 56 sequences were aligned to each other and to the 28 sequences derived from amplification mediated by primers p1, p2, and p4 combined with primers p7 or p8 (described above). Figure 7 shows that there were 26 distinct sequences. The average pairwise distance between P. murina MSG gene sequences was 7% (range: 1–16%). The hypothesis that the gene family contains 26 members fit well with the results of real-time PCR experiments on the CRJE described above (Figure 5B).
Figure 7. Neighbor-joining tree of MSG sequences.
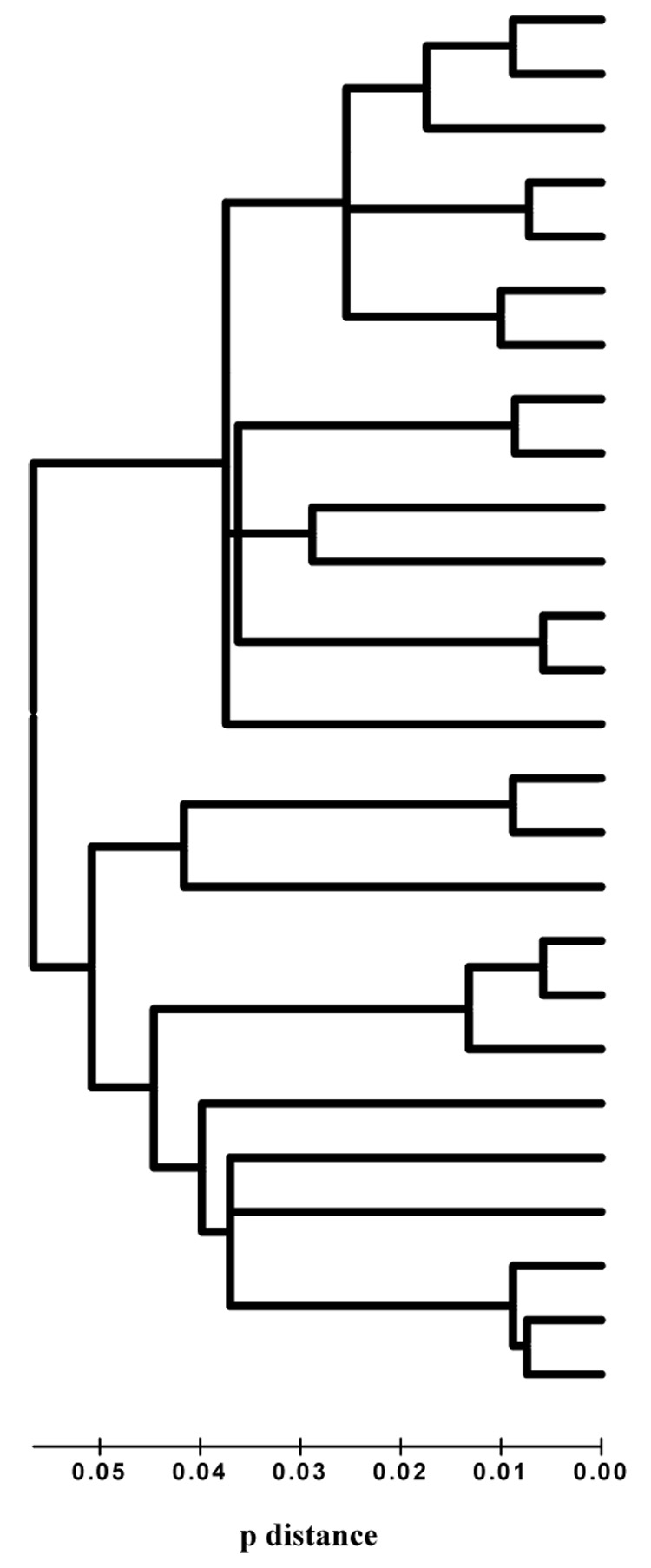
Branch lengths are drawn to scale. Sequences less than 1% diverged were placed on the same branch. The scale at the bottom shows branch lengths corresponding to pairwise p-distance values calculated by dividing the number of mismatched bases by the total number compared.
Alignment also showed that the sequences downstream of primer p3.1 were nearly invariant for 132 bp (Figure 6), suggesting that the P. murina CRJE is 132 bp long.
While the P. murina CRJE was strongly conserved, this element was not invariant. For example, four cloned copies of this sequence contained three tandem copies of the 12 bp sequence 5’:CAAGCAGCAGGA: 3’, which was present only twice in the other 80 copies sequenced (Figure 6). Repeat expansion can occur during PCR, but expansion is much less frequent than contraction (Shinde, Deepali et al. 2003). Therefore, it would appear that the predominant CRJE allele has two copies of the 12 bp sequence, and that the 3-copy allele is a natural variant rather than a PCR artifact. Variation of this sort has been seen in the expression sites of other species of Pneumocystis (Sunkin, S. M. and Stringer, J. R. 1996; Ma, L. et al. 2002). Transition mutations were also observed within the P. murina CRJE, but these were very rare, generally occurring in only one sequence out of 84. PCR error can cause variation at this frequency (Tindall, K. R. and Kunkel, T. A. 1988).
Comparison of expression sites in different Pneumocystis species
Figure 8 summarizes the current picture of expression-site structure in four species of Pneumocystis. In P. carinii, all but the last 28 bp of the UCS (i.e. the sequence found at the beginning of all mRNAs encoding an MSG) are encoded by DNA that is unique in the genome. The last 28 bp of the UCS are encoded by a copy of the CRJE, other copies of which occur at the beginning of MSG genes that are not at the expression site. By contrast, in P. murina, all but the last 132 bp of the UCS are encoded by DNA that is unique in the genome. Thus, the CRJE in P. murina appears to be much larger than it is in P. carinii. It is possible that the P. murina CRJE will shrink when more P. murina MSG genes are sequenced. However, quantitative PCR analysis of the central 63 basepairs of the CRJE indicated that the number of MSG genes in P. murina is approximately the same as the number of different MSG sequences observed so far, suggesting that the CRJE size estimate is based on the vast majority of the gene family members in this species.
Figure 8. Comparison of expression sites in four Pneumocystis species.
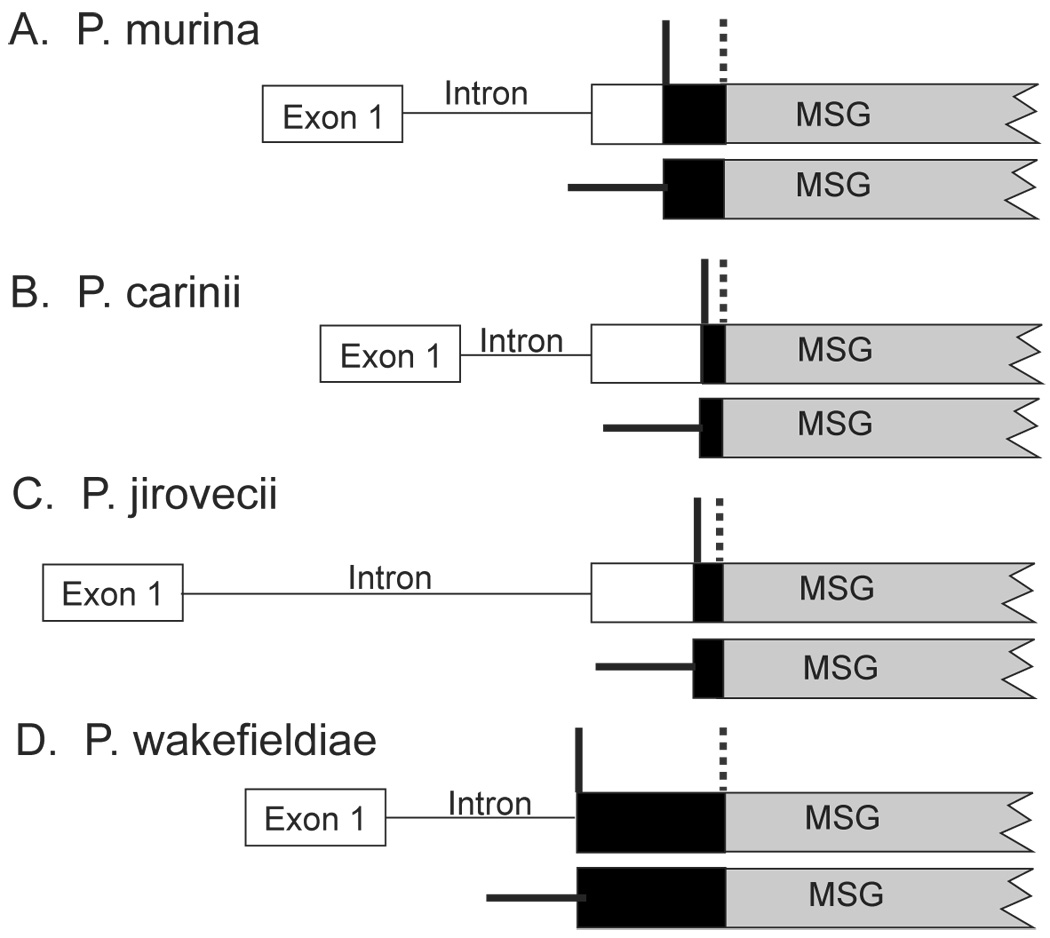
For each species, two maps are shown. The upper map depicts the expresson site and the beginning of an expression-site-linked MSG gene. The lower map depicts an MSG gene not linked to the expression site. The symbol │ indicates the boundary between unique and repeated DNA at the expression site. The region to the left of the line contains DNA that is present only once in the haploid genome of P. murina. The region to the right of the line contains DNA that is a member of a family of sequences located throughout the genome. The symbol  marks the boundary between the UCS and MSG sequences. The black boxes represent CRJEs, defined as the 5-prime sequence that is conserved among all members of the MSG gene family in a given species.
marks the boundary between the UCS and MSG sequences. The black boxes represent CRJEs, defined as the 5-prime sequence that is conserved among all members of the MSG gene family in a given species.
The expression sites of P. jirovecii and P. wakefieldiae have not been as thoroughly studied as those in P. murina and P. carinii, but current data suggest that these two species also are each distinct in this respect. In P. jirovecii, DNA hybridization data have indicated that nearly all of the DNA encoding the UCS is located solely at the expression-site (Kutty, G., Ma, L., and Kovacs, J. A. 2001). To examine this issue at the sequence level, we aligned the sequence at the P. jirovecii expression site (Kutty, G., Ma, L., and Kovacs, J. A. 2001) to the sequences upstream of three P. jirovecii MSG genes (Garbe, T. R. and Stringer, J. R. 1994). The first 18 nucleotides upstream of the sequence homologous to the 28 bp P. carinii CRJE were present at both the expression site and in regions upstream of MSG genes not at the expression site (data not shown). These data suggest that the CRJE in P. jirovecii may be as large as 46 bp. However, analysis of regions upstream of additional MSG genes may show that the CRJE in P. jirovecii is smaller than it now appears. In any event, the P. jirovecii CRJE appears to be substantially smaller than the P. murina CRJE. The expression-site locus of P. wakefieldiae appears to be quite different from those in the other three species. In P. wakefieldiae, more than half of the UCS is encoded by DNA that is repeated in the genome (Schaffzin, J. K. and Stringer, J. R. 2000).
Discussion
Comparison of the UCS locus and attached MSG genes in four species of Pneumocystis showed that the structure of the expression site is conserved across species, but no two species are identical in this regard. For example, each species has an intron in the unique DNA encoding the UCS, and this intron is exactly the same distance from the beginning of the UCS open reading frame in all four species. However, the introns of different species are different in length and sequence. A second example is the CRJE. All four species have a sequence nearly identical to the 28 bp CRJE of P. carinii. However, the P. carinii CRJE appears to be rather small compared to the 132 bp CRJE of P. murina. The CRJE of P. murina is also much larger than the CRJEs in P. jirovecii. The P. wakefieldiae CRJE appears to be larger still, at over 300 bp in length (Schaffzin, J. K. and Stringer, J. R. 2000).
CRJE size differences may have functional consequences. It is possible that the CRJE serves to mediate recombination events that switch the MSG sequence that is at the expression site. If such events were to be due to the action of a site-specific recombinase, then it would seem that each species may have evolved its own such enzyme. A very large CRJE, such that in P. wakefieldiae, might allow homologous recombination between CRJE copies to contribute to variation at the expression site. Sequences 300 bp in length support efficient homologous recombination in Saccharomyces cerevisiae (Jinks-Robertson, S., Michelitch, M., and Ramcharan, S. 1993).
While the mechanism that generates change at the expression site is not completely understood, such a mechanism appears to be at work in P. murina because multiple MSG genes were observed at the expression-site locus. This situation is similar to that observed in other Pneumocystis species (Wada, M. et al. 1995; Sunkin, S. M. and Stringer, J. R. 1996; Sunkin, S. M. and Stringer, J. R. 1997; Schaffzin, J. K. and Stringer, J. R. 2000; Kutty, G., Ma, L., and Kovacs, J. A. 2001; Keely, S. P., Cushion, M. T., and Stringer, J. R. 2003). Studying the switching process has been difficult because Pneumocystis does not proliferate much in culture. Progress in understanding the switching process would be facilitated by developing assays utilizing animals infected with a small number of Pneumocystis expressing one, known MSG gene. Under these conditions, organisms expressing a different MSG are expected to emerge as the population expands in the animal. The rate of appearance of these “switched” organisms would provide an indication of the frequency of switching, and the structure of the expression-site linked MSG gene before and after switching would indicate the mechanism of switching. Studies on switching in P. carinii were initially promising, but have been hampered by the tendency of laboratory rats to be colonized by this fungus (Keely, S. P., Cushion, M. T., and Stringer, J. R. 2003). Such animals are difficult to use to study the frequency and mechanism of switching. By contrast, laboratory mice free of P. murina are routinely available. Therefore, it should be easier to acquire information on switching by studying P. murina.
The ability to switch MSG genes at the expression-site locus is a means to generate surface variability, which is probably an important feature of the Pneumocystis lifestyle. Programmed surface variation mediated by differential expression of gene families is common among microbes that must avoid the host immune response in order to survive (Borst, P. 2002; Recker, M. et al. 2004; Gatton, M. L. and Cheng, Q. 2004; Andrews, T. D. and Gojobori, T. 2004; Embers, M. E., Ramamoorthy, R., and Philipp, M. T. 2004; Liang, F. T. et al. 2004). Most examples of antigenic variation come from highly virulent microbes such as African trypanosomes, which use antigenic variation to maintain a large population in the blood stream despite a strong host immune response (Vickerman, K. 1978). By contrast, P. murina is unable to cause disease in immunocompetent mice, but is able to cause transient infections in them (Gigliotti, F., Harmsen, A. G., and Wright, T. W. 2003; An, C. L., Gigliotti, F., and Harmsen, A. G. 2003; Chabe, M. et al. 2004). Disease is seen only when the immune system is debilitated. The same seems to be true in other host species, including wild and laboratory animals, and humans (Mazars, E. et al. 1997; Bishop, R. et al. 1997; Nielsen, M. H. et al. 1998; Laakkonen, J. 1998; Weisbroth, S. H. et al. 1999; Denis, C. M. et al. 2000; Palmer, R. J. et al. 2000; Vargas, S. L. et al. 2001; Demanche, C. et al. 2001; Icenhour, C. R. et al. 2001; Durand-Joly, I. et al. 2003). Therefore, the function of the MSG system is probably not to overcome the immune response and cause disease. Instead, MSG variation might be used to prolong colonization by producing cells that are not recognized by the defense mechanisms deployed against cells with the most common antigenic phenotype.
The frequent-colonization hypothesis fits with the observation that each host species has its own kind of Pneumocystis, suggesting that the microbe and host have coevolved, and that each species of Pneumocystis depends on a particular host species for survival (Guillot, J. et al. 2001). This suggestion is supported by the fastidiousness of these microbes. They do not thrive in culture, and seem to be able to propagate continuously only in the lungs of the host in which they are found (Gigliotti, F. et al. 1993; Wakefield, A. E. et al. 1998; Dei-Cas, E. 2000; Durand-Joly, I. et al. 2002). It is reasonable to speculate that the P. murina MSG system serves to foster survival of relatively low numbers of P. murina in the lungs of immunocompetent mice, and that this survival is necessary to assure transmission to other mice.
Acknowledgements
This work was supported by grant: R01AI36701 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
LITERATURE CITED
- An CL, Gigliotti F, Harmsen AG. Exposure of immunocompetent adult mice to Pneumocystis carinii f. sp. muris by cohousing: growth of P. carinii f. sp. muris and host immune response. Infect. Immun. 2003;71:2065–2070. doi: 10.1128/IAI.71.4.2065-2070.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews TD, Gojobori T. Strong positive selection and recombination drive the antigenic variation of the PilE protein of the human pathogen Neisseria meningitidis. Genetics. 2004;166:25–32. doi: 10.1534/genetics.166.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angus CW, Tu A, Vogel P, Qin M, Kovacs JA. Expression of variants of the major surface glycoprotein of Pneumocystis carinii. J. Exp. Med. 1996;183:1229–1234. doi: 10.1084/jem.183.3.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong MY, Cushion MT. Animal Models. In: Walzer PD, editor. Pneumocystis carinii pneumonia. 2nd. New York: Marcel Dekker; 1994. pp. 181–222. [Google Scholar]
- Bishop R, Gurnell J, Laakkonen J, Whitwell K, Peters S. Detection of Pneumocystis DNA in the lungs of several species of wild mammal. J. Eukaryot. Microbiol. 1997;44:57S. doi: 10.1111/j.1550-7408.1997.tb05777.x. [DOI] [PubMed] [Google Scholar]
- Borst P. Antigenic variation and allelic exclusion. Cell. 2002;109:5–8. doi: 10.1016/s0092-8674(02)00711-0. [DOI] [PubMed] [Google Scholar]
- Chabe M, Dei-Cas E, Creusy C, Fleurisse L, Respaldiza N, Camus D, Durand-Joly I. Immunocompetent hosts as a reservoir of pneumocystis organisms: histological and rt-PCR data demonstrate active replication. Eur. J. Clin. Microbiol Infect. Dis. 2004;23:89–97. doi: 10.1007/s10096-003-1092-2. [DOI] [PubMed] [Google Scholar]
- Cornillot E, Keller B, Cushion MT, Metenier G, Vivares CP. Fine analysis of the Pneumocystis carinii f. sp. carinii genome by two- dimensional pulsed-field gel electrophoresis. Gene. 2002;293:87–95. doi: 10.1016/s0378-1119(02)00604-2. [DOI] [PubMed] [Google Scholar]
- Cushion MT, Kaselis M, Stringer SL, Stringer JR. Genetic stability and diversity of Pneumocystis carinii infecting rat colonies. Infect. Immun. 1993;61:4801–4813. doi: 10.1128/iai.61.11.4801-4813.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushion MT, Orr S, Keely SP, Stringer JR. Time between inoculations and karyotype forms of Pneumocystis carinii f. sp. carinii influence outcome of experimental coinfections in rats. Infect. Immun. 2001;69:97–107. doi: 10.1128/IAI.69.1.97-107.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dei-Cas E. Pneumocystis infections: the iceberg? Med. Mycol. 2000;38 Suppl 1:23–32. [PubMed] [Google Scholar]
- Dei-Cas E, Brun-Pascaud M, Bille-Hansen V, Allaert A, Aliouat EM. Animal models of pneumocystosis. FEMS Immunol. Med. Microbiol. 1998;22:163–168. doi: 10.1111/j.1574-695X.1998.tb01201.x. [DOI] [PubMed] [Google Scholar]
- Demanche C, Berthelemy M, Petit T, Polack B, Wakefield AE, Dei-Cas E, Guillot J. Phylogeny of Pneumocystis carinii from 18 primate species confirms host specificity and suggests coevolution. J. Clin. Microbiol. 2001;39:2126–2133. doi: 10.1128/JCM.39.6.2126-2133.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis CM, Mazars E, Guyot K, Odberg-Ferragut C, Viscogliosi E, Dei-Cas E, Wakefield AE. Genetic divergence at the SODA locus of six different formae speciales of Pneumocystis carinii. Med. Mycol. 2000;38:289–300. doi: 10.1080/mmy.38.4.289.300. [DOI] [PubMed] [Google Scholar]
- Durand-Joly I, Aliouat eM, Recourt C, Guyot K, Francois N, Wauquier M, Camus D, Dei-Cas E. Pneumocystis carinii f. sp. hominis is not infectious for SCID mice. J. Clin. Microbiol. 2002;40:1862–1865. doi: 10.1128/JCM.40.5.1862-1865.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand-Joly I, Soula F, Chabe M, Dalle JH, Lafitte JJ, Senechal M, Pinon A, Camus D, Dei-Cas E. Long-term colonization with Pneumocystis jirovecii in hospital staffs: a challenge to prevent nosocomial pneumocystosis. J. Eukaryot. Microbiol. 2003;50 Suppl:614–615. doi: 10.1111/j.1550-7408.2003.tb00650.x. [DOI] [PubMed] [Google Scholar]
- Edman JC, Hatton TW, Nam M, Turner R, Mei Q, Angus CW, Kovacs JA. A single expression site with a conserved leader sequence regulates variation of expression of the Pneumocystis carinii family of major surface glycoprotein genes. DNA Cell Biol. 1996;15:989–999. doi: 10.1089/dna.1996.15.989. [DOI] [PubMed] [Google Scholar]
- Embers ME, Ramamoorthy R, Philipp MT. Survival strategies of Borrelia burgdorferi, the etiologic agent of Lyme disease. Microbes. Infect. 2004;6:312–318. doi: 10.1016/j.micinf.2003.11.014. [DOI] [PubMed] [Google Scholar]
- Empey KM, Hollifield M, Schuer K, Gigliotti F, Garvy BA. Passive immunization of neonatal mice against Pneumocystis carinii f. sp. muris enhances control of infection without stimulating inflammation. Infect. Immun. 2004;72:6211–6220. doi: 10.1128/IAI.72.11.6211-6220.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel JK. Pneumocystis jiroveci n. sp. from man: morphology, physiology, and immunology in relation to pathology. National. Cancer Institute. Monographs. 1976;43:13–30. [PubMed] [Google Scholar]
- Frenkel JK. Pneumocystis pneumonia, an immunodeficiency-dependent disease (IDD): a critical historical overview. J. Eukaryot. Microbiol. 1999;46:89S–92S. [PubMed] [Google Scholar]
- Garbe TR, Stringer JR. Molecular characterization of clustered variants of genes encoding major surface antigens of human Pneumocystis carinii. Infect. Immun. 1994;62:3092–3101. doi: 10.1128/iai.62.8.3092-3101.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatton ML, Cheng Q. Investigating antigenic variation and other parasite-host interactions in Plasmodium falciparum infections in naive hosts. Parasitology. 2004;128:367–376. doi: 10.1017/s0031182003004608. [DOI] [PubMed] [Google Scholar]
- Gigliotti F. Host species-specific antigenic variation of a mannosylated surface glycoprotein of Pneumocystis carinii. J. Infect. Dis. 1992;165:329–336. doi: 10.1093/infdis/165.2.329. [DOI] [PubMed] [Google Scholar]
- Gigliotti F, Harmsen AG, Haidaris CG, Haidaris PJ. Pneumocystis carinii is not universally transmissible between mammalian species. Infect. Immun. 1993;61:2886–2890. doi: 10.1128/iai.61.7.2886-2890.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigliotti F, Harmsen AG, Wright TW. Characterization of transmission of Pneumocystis carinii f. sp. muris through immunocompetent BALB/c mice. Infect. Immun. 2003;71:3852–3856. doi: 10.1128/IAI.71.7.3852-3856.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillot J, Demanche C, Hugot JP, Berthelemy M, Wakefield AE, Dei-Cas E, Chermette R. Parallel phylogenies of Pneumocystis species and their mammalian hosts. J. Eukaryot. Microbiol. 2001 Suppl:113S–115S. doi: 10.1111/j.1550-7408.2001.tb00475.x. [DOI] [PubMed] [Google Scholar]
- Haidaris CG, Medzihradsky OF, Gigliotti F, Simpson-Haidaris PJ. Molecular characterization of mouse Pneumocystis carinii surface glycoprotein A. DNA Res. 1998;5:77–85. doi: 10.1093/dnares/5.2.77. [DOI] [PubMed] [Google Scholar]
- Haidaris CG, Wright TW, Gigliotti F, Haidaris PJ. Molecular cloning and characterization of ferret Pneumocystis carinii gp120. J. Protozool. 1991;38:5S–6S. [PubMed] [Google Scholar]
- Haidaris PJ, Wright TW, Gigliotti F, Haidaris CG. Expression and characterization of a cDNA clone encoding an immunodominant surface glycoprotein of Pneumocystis carinii. J. Infect. Dis. 1992;166:1113–1123. doi: 10.1093/infdis/166.5.1113. [DOI] [PubMed] [Google Scholar]
- Hong ST, Steele PE, Cushion MT, Walzer PD, Stringer SL, Stringer JR. Pneumocystis carinii karyotypes. J. Clin. Microbiol. 1990;28:1785–1795. doi: 10.1128/jcm.28.8.1785-1795.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Icenhour CR, Rebholz SL, Collins MS, Cushion MT. Widespread Occurrence of Pneumocystis carinii in Commercial Rat Colonies Detected Using Targeted PCR and Oral Swabs. J. Clin. Microbiol. 2001;39:3437–3441. doi: 10.1128/JCM.39.10.3437-3441.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinks-Robertson S, Michelitch M, Ramcharan S. Substrate length requirements for efficient mitotic recombination in Saccharomyces cerevisiae. Mol. Cell Biol. 1993;13:3937–3950. doi: 10.1128/mcb.13.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson AB, Nyberg G, Normark S. Phase variation of gonococcal pili by frameshift mutation in pilC, a novel gene for pilus assembly. EMBO J. 1991;10:477–488. doi: 10.1002/j.1460-2075.1991.tb07970.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keely SP, Cushion MT, Stringer JR. Diversity at the Locus Associated with Transcription of a Variable Surface Antigen of Pneumocystis carinii as an Index of Population Structure and Dynamics in Infected Rats. Infect. Immun. 2003;71:47–60. doi: 10.1128/IAI.71.1.47-60.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keely SP, Fischer JM, Cushion MT, Stringer JR. Phylogenetic identification of Pneumocystis murina sp. nov., a new species in laboratory mice. Microbiology. 2004;150:1153–1165. doi: 10.1099/mic.0.26921-0. [DOI] [PubMed] [Google Scholar]
- Keely SP, Renauld H, Wakefield AE, Cushion MT, Smulian AG, Fosker N, Fraser A, Harris D, Murphy L, Price C, Quail MA, Seeger K, Sharp S, Tindal CJ, Warren T, Zuiderwijk E, Barrell BG, Stringer JR, Hall N. Gene arrays at Pneumocystis carinii telomeres. Genetics. 2005;170:1589–1600. doi: 10.1534/genetics.105.040733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keely SP, Stringer JR. Nomenclature and genetic variation of Pneumocystis. In: Walzer PD, Cushion MT, editors. Pneumocystis Pneumonia. 3. New York: Marcel Dekker; 2005. pp. 39–59. [Google Scholar]
- Kitada K, Wada M, Nakamura Y. Multi-gene family of major surface glycoproteins of Pneumocystis carinii: full-size cDNA cloning and expression. DNA Res. 1994;1:57–66. doi: 10.1093/dnares/1.2.57. [DOI] [PubMed] [Google Scholar]
- Kovacs JA, Powell F, Edman JC, Lundgren B, Martinez A, Drew B, Angus CW. Multiple genes encode the major surface glycoprotein of Pneumocystis carinii. J. Biol. Chem. 1993;268:6034–6040. [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Kutty G, Ma L, Kovacs JA. Characterization of the expression site of the major surface glycoprotein of human-derived Pneumocystis carinii. Mol. Microbiol. 2001;42:183–193. doi: 10.1046/j.1365-2958.2001.02620.x. [DOI] [PubMed] [Google Scholar]
- Laakkonen J. Pneumocystis carinii in wildlife. Int. J. Parasitol. 1998;28:241–252. doi: 10.1016/s0020-7519(97)00155-0. [DOI] [PubMed] [Google Scholar]
- Larsen HH, Kovacs JA, Stock F, Vestereng VH, Lundgren B, Fischer SH, Gill VJ. Development of a rapid real-time PCR assay for quantitation of Pneumocystis carinii f. sp. carinii. J. Clin. Microbiol. 2002;40:2989–2993. doi: 10.1128/JCM.40.8.2989-2993.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LH, Gigliotti F, Wright TW, Simpson-Haidaris PJ, Weinberg GA, Haidaris CG. Molecular characterization of KEX1, a kexin-like protease in mouse Pneumocystis carinii. Gene. 2000;242:141–150. doi: 10.1016/s0378-1119(99)00533-8. [DOI] [PubMed] [Google Scholar]
- Liang FT, Yan J, Mbow ML, Sviat SL, Gilmore RD, Mamula M, Fikrig E. Borrelia burgdorferi changes its surface antigenic expression in response to host immune responses. Infect. Immun. 2004;72:5759–5767. doi: 10.1128/IAI.72.10.5759-5767.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke M, Ashbaugh A, Demland J, Koch J, Tanaka R, Walzer P. Resolution of Pneumocystis murina infection following withdrawal of corticosteroid induced immunosuppression. Microb. Pathog. 2006a;40:15–22. doi: 10.1016/j.micpath.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Linke M, Ashbaugh A, Koch J, Tanaka R, Walzer P. Surfactant protein A limits Pneumocystis murina infection in immunosuppressed C3H/HeN mice and modulates host response during infection. Microbes. Infect. 2005;7:748–759. doi: 10.1016/j.micinf.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Linke M, Ashbaugh A, Koch J, Tanaka R, Walzer P. Efficient resolution of Pneumocystis murina infection in surfactant protein A-deficient mice following withdrawal of corticosteroid-induced immunosuppression. J. Med. Microbiol. 2006b;55:143–147. doi: 10.1099/jmm.0.46190-0. [DOI] [PubMed] [Google Scholar]
- Linke MJ, Cushion MT, Walzer PD. Properties of the major antigens of rat and human Pneumocystis carinii. Infect. Immun. 1989;57:1547–1555. doi: 10.1128/iai.57.5.1547-1555.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund FE, Schuer K, Hollifield M, Randall TD, Garvy BA. Clearance of Pneumocystis carinii in mice is dependent on B cells but not on P. carinii-specific antibody. J. Immunol. 2003;171:1423–1430. doi: 10.4049/jimmunol.171.3.1423. [DOI] [PubMed] [Google Scholar]
- Ma L, Kutty G, Jia Q, Imamichi H, Huang L, Atzori C, Beckers P, Groner G, Beard CB, Kovacs JA. Analysis of variation in tandem repeats in the intron of the major surface glycoprotein expression site of the human form of Pneumocystis carinii. J. Infect. Dis. 2002;186:1647–1654. doi: 10.1086/345721. [DOI] [PubMed] [Google Scholar]
- Mazars E, Guyot K, Fourmaintraux S, Renaud F, Petavy F, Camus D, Dei-Cas E. Detection of Pneumocystis in European wild animals. J. Eukaryot. Microbiol. 1997;44:39S. doi: 10.1111/j.1550-7408.1997.tb05763.x. [DOI] [PubMed] [Google Scholar]
- McAllister F, Steele C, Zheng M, Young E, Shellito JE, Marrero L, Kolls JK. T cytotoxic-1 CD8+ T cells are effector cells against pneumocystis in mice. J. Immunol. 2004;172:1132–1138. doi: 10.4049/jimmunol.172.2.1132. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Kitada K, Wada M, Saito M. Epitope study and cDNA screening of major surface glycoprotein of Pneumocystis carinii. J. Protozool. 1991;38:3S–4S. [PubMed] [Google Scholar]
- Nielsen MH, Settnes OP, Aliouat EM, Cailliez JC, Dei-Cas E. Different ultrastructural morphology of Pneumocystis carinii derived from mice, rats, and rabbits. APMIS. 1998;106:771–779. [PubMed] [Google Scholar]
- Palmer RJ, Settnes OP, Lodal J, Wakefield AE. Population structure of rat-derived Pneumocystis carinii in Danish wild rats. Appl. Environ. Microbiol. 2000;66:4954–4961. doi: 10.1128/aem.66.11.4954-4961.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi MH, Empey KM, Garvy BA. Modulation of proinflammatory responses to Pneumocystis carinii f. sp. muris in neonatal mice by granulocyte-macrophage colony-stimulating factor and IL-4: role of APCs. J. Immunol. 2005;174:441–448. doi: 10.4049/jimmunol.174.1.441. [DOI] [PubMed] [Google Scholar]
- Qureshi MH, Harmsen AG, Garvy BA. IL-10 modulates host responses and lung damage induced by Pneumocystis carinii infection. J. Immunol. 2003;170:1002–1009. doi: 10.4049/jimmunol.170.2.1002. [DOI] [PubMed] [Google Scholar]
- Recker M, Nee S, Bull PC, Kinyanjui S, Marsh K, Newbold C, Gupta S. Transient cross-reactive immune responses can orchestrate antigenic variation in malaria. Nature. 2004;429:555–558. doi: 10.1038/nature02486. [DOI] [PubMed] [Google Scholar]
- Redhead SA, Cushion MT, Frenkel JK, Stringer JR. Pneumocystis and Trypanosoma cruzi: Nomenclature and Typifications. J. Eukaryot. Microbiol. 2006;53:2–11. doi: 10.1111/j.1550-7408.2005.00072.x. [DOI] [PubMed] [Google Scholar]
- Sambrook JF, Fristch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd 1989. [Google Scholar]
- Schaffzin JK, Garbe TR, Stringer JR. Major surface glycoprotein genes from pneumocystis carinii f. sp. ratti. Fungal. Genet. Biol. 1999;28:214–226. doi: 10.1006/fgbi.1999.1171. [DOI] [PubMed] [Google Scholar]
- Schaffzin JK, Stringer JR. The major surface glycoprotein expression sites of two special forms of rat Pneumocystis carinii differ in structure. J. Infect. Dis. 2000;181:1729–1739. doi: 10.1086/315438. [DOI] [PubMed] [Google Scholar]
- Schaffzin JK, Stringer JR. Expression of the Pneumocystis carinii major surface glycoprotein epitope is correlated with linkage of the cognate gene to the upstream conserved sequence locus. Microbiology. 2004;150:677–686. doi: 10.1099/mic.0.26542-0. [DOI] [PubMed] [Google Scholar]
- Shinde D, Lai Y, Sun F, Arnheim N. Taq DNA polymerase slippage mutation rates measured by PCR and quasi-likelihood analysis: (CA/GT)n and (A/T)n microsatellites. Nucl. Acids Res. 2003;31:974–980. doi: 10.1093/nar/gkg178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer JR. The MSG Gene Family and Antigenic Variation in the Fungus P. carinii. In: Craig AG, Scherf A, editors. Antigenic Variation. 1. New York: Academic Press; 2003. pp. 100–120. [Google Scholar]
- Stringer JR. Surface Antigens. In: Walzer PD, Cushion MT, editors. Pneumocystis Pneumonia. 3. New York: Marcel Dekker; 2005. pp. 95–126. [Google Scholar]
- Stringer JR, Beard CB, Miller RF, Wakefield AE. A New Name (Pneumocystis jiroveci) for Pneumocystis from Humans. Emerg. Infect. Dis. 2002;8:891–896. doi: 10.3201/eid0809.020096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer JR, Cushion MT, Wakefield AE. New nomenclature for the genus Pneumocystis; Journal of Eukaryotic Microbiology, Supplement:Proceedings of the Seventh International Workshops on Opportunistic Protists; 2001. pp. 184s–189s. [DOI] [PubMed] [Google Scholar]
- Stringer JR, Keely SP. Genetics of surface antigen expression in Pneumocystis carinii. Infect. Immun. 2001;69:627–639. doi: 10.1128/IAI.69.2.627-639.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer SL, Garbe T, Sunkin SM, Stringer JR. Genes encoding antigenic surface glycoproteins in Pneumocystis from humans. J. Eukaryot. Microbiol. 1993;40:821–826. doi: 10.1111/j.1550-7408.1993.tb04481.x. [DOI] [PubMed] [Google Scholar]
- Stringer SL, Hong ST, Giuntoli D, Stringer JR. Repeated DNA in Pneumocystis carinii. J. Clin. Microbiol. 1991;29:1194–1201. doi: 10.1128/jcm.29.6.1194-1201.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkin SM, Linke MJ, McCormack FX, Walzer PD, Stringer JR. Identification of a putative precursor to the major surface glycoprotein of Pneumocystis carinii. Infect. Immun. 1998;66:741–746. doi: 10.1128/iai.66.2.741-746.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkin SM, Stringer JR. Translocation of surface antigen genes to a unique telomeric expression site in Pneumocystis carinii. Mol. Microbiol. 1996;19:283–295. doi: 10.1046/j.1365-2958.1996.375905.x. [DOI] [PubMed] [Google Scholar]
- Sunkin SM, Stringer JR. Residence at the expression site is necessary and sufficient for the transcription of surface antigen genes of Pneumocystis carinii. Molecular Microbiology. 1997;25:147–160. doi: 10.1046/j.1365-2958.1997.4461806.x. [DOI] [PubMed] [Google Scholar]
- Sunkin SM, Stringer SL, Stringer JR. A tandem repeat of rat-derived Pneumocystis carinii genes encoding the major surface glycoprotein. J. Eukaryot. Microbiol. 1994;41:292–300. doi: 10.1111/j.1550-7408.1994.tb01509.x. [DOI] [PubMed] [Google Scholar]
- Tanabe K, Takasaki S, Watanabe J, Kobata A, Egawa K, Nakamura Y. Glycoproteins composed of major surface immunodeterminants of Pneumocystis carinii. Infect. Immun. 1989;57:1363–1368. doi: 10.1128/iai.57.5.1363-1368.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CF, Jr, Limper AH. Pneumocystis pneumonia. N. Engl. J. Med. 2004;350:2487–2498. doi: 10.1056/NEJMra032588. [DOI] [PubMed] [Google Scholar]
- Tindall KR, Kunkel TA. Fidelity of DNA synthesis by the Thermus aquaticus DNA polymerase. Biochemistry. 1988;27:6008–6013. doi: 10.1021/bi00416a027. [DOI] [PubMed] [Google Scholar]
- Vargas SL, Hughes WT, Santolaya ME, Ulloa AV, Ponce CA, Cabrera CE, Cumsille F, Gigliotti F. Search for Primary Infection by Pneumocystis carinii in a Cohort of Normal, Healthy Infants. Clin. Infect. Dis. 2001;32:855–861. doi: 10.1086/319340. [DOI] [PubMed] [Google Scholar]
- Vickerman K. Antigenic variation in trypanosomes. Nature. 1978;273:613–617. doi: 10.1038/273613a0. [DOI] [PubMed] [Google Scholar]
- Wada M, Kitada K, Saito M, Egawa K, Nakamura Y. cDNA sequence diversity and genomic clusters of major surface glycoprotein genes of Pneumocystis carinii. J. Infect. Dis. 1993;168:979–985. doi: 10.1093/infdis/168.4.979. [DOI] [PubMed] [Google Scholar]
- Wada M, Nakamura Y. MSG gene cluster encoding major cell surface glycoproteins of rat Pneumocystis carinii. DNA Res. 1994;1:163–168. doi: 10.1093/dnares/1.4.163. [DOI] [PubMed] [Google Scholar]
- Wada M, Nakamura Y. Unique telomeric expression site of major-surface-glycoprotein genes of Pneumocystis carinii. DNA Res. 1996;3:55–64. doi: 10.1093/dnares/3.2.55. [DOI] [PubMed] [Google Scholar]
- Wada M, Sunkin SM, Stringer JR, Nakamura Y. Antigenic variation by positional control of major surface glycoprotein gene expression in Pneumocystis carinii. J. Infect. Dis. 1995;171:1563–1568. doi: 10.1093/infdis/171.6.1563. [DOI] [PubMed] [Google Scholar]
- Wakefield AE, Stringer JR, Tamburrini E, Dei-Cas E. Genetics, metabolism and host specificity of Pneumocystis carinii. Med. Mycol. 1998;36 Suppl 1:183–193. [PubMed] [Google Scholar]
- Walzer PD, Ashbaugh A, Collins M, Cushion MT. Anti-human immunodeficiency virus drugs are ineffective against Pneumocystis carinii in vitro and in vivo. J. Infect. Dis. 2001;184:1355–1357. doi: 10.1086/323991. [DOI] [PubMed] [Google Scholar]
- Weisbroth SH, Geistfeld J, Weisbroth SP, Williams B, Feldman SH, Linke MJ, Orr S, Cushion MT. Latent pneumocystis carinii infection in commercial rat colonies: comparison of inductive immunosuppressants plus histopathology, PCR, and serology as detection methods. J. Clin. Microbiol. 1999;37:1441–1446. doi: 10.1128/jcm.37.5.1441-1446.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright TW, Gigliotti F, Haidaris CG, Simpson-Haidaris PJ. Cloning and characterization of a conserved region of human and rhesus macaque Pneumocystis carinii gpA. Gene. 1995;167:185–189. doi: 10.1016/0378-1119(95)00704-0. [DOI] [PubMed] [Google Scholar]
- Wright TW, Notter RH, Wang Z, Harmsen AG, Gigliotti F. Pulmonary inflammation disrupts surfactant function during Pneumocystis carinii pneumonia. Infect. Immun. 2001;69:758–764. doi: 10.1128/IAI.69.2.758-764.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright TW, Simpson Haidaris PJ, Gigliotti F, Harmsen AG, Haidaris CG. Conserved sequence homology of cysteine-rich regions in genes encoding glycoprotein A in Pneumocystis carinii derived from different host species. Infect. Immun. 1994;62:1513–1519. doi: 10.1128/iai.62.5.1513-1519.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyder MA, Rasch EM, Kaneshiro ES. Quantitation of absolute Pneumocystis carinii nuclear DNA content. Trophic and cystic forms isolated from infected rat lungs are haploid organisms. J. Eukaryot. Microbiol. 1998;45:233–239. doi: 10.1111/j.1550-7408.1998.tb04531.x. [DOI] [PubMed] [Google Scholar]


