Domestic cats (Felis silvestris catus) (herein referred to as “cats”) are neither attracted to, nor show avoidance of the taste of sweet carbohydrates and high-intensity sweeteners (1-3), yet they do show a preference for selected amino acids (4), and avoid stimuli that taste either bitter or very sour to humans (1,4). Consistent with this behavioral evidence, recordings from cat taste nerve fibers and from units of the geniculate ganglion innervating taste cells demonstrated responses to salty, sour, and bitter stimuli as well as to amino acids and nucleotides, but showed no response to sucrose and several other sugars (4-11). The sense of taste in cats appears similar to that of other mammals with the exception of an inability to taste sweet stimuli.
Because only the sweet taste modality appears absent, we postulated that the defect in cats (and likely in other obligate carnivores of Felidae) lay at the receptor step, subtending this modality. The possible defects at the molecular level could range from a single to a few amino acid substitutions, such as is found between sweet “taster” and “nontaster” strains of mice (12-14), to more radical mechanisms, such as an unexpressed pseudogene.
To distinguish among these possibilities, we identified the DNA sequences and examined the structures of the 2 known genes Tas1r2 and Tas1r3 that encode the sweet taste receptor heteromer T1R2/T1R3 in other mammals. We compared these with the sequence and structure of the same genes in dogs, humans, mice and rats, all species that respond to sweet stimuli.
Molecular cloning of cat Tas1r3 and Tas1r2
We identified 2 receptor genes, Tas1r3 and Tas1r2, in domestic cats by screening a feline genomic BAC library and performing PCR with degenerate primers on cat genomic DNA. Using the same strategy as for the canine genomic BAC library, we also identified the same 2 genes from dogs.
The cat Tas1r3 gene shows high similarity with those of dogs, humans, mice and rats at both the cDNA (from 74 to 87%) and deduced amino acid level (from 72 to 85%). To confirm the exon-intron boundaries for cat Tas1r3, we performed both RT-PCR on cDNA from cat taste bud-containing circumvallate and fungiform papillae and PCR on cat genomic DNA using intron spanning primers, and compared the cDNA sequence with the genomic sequence (data not shown). Both the cat Tas1r3 and dog Tas1r3 genes are composed of 6 similarly sized exons and 5 introns (Fig. 1a). There was nothing within the cat Tas1r3 gene that would suggest that the cat gene was defective compared with that of the dog.
FIGURE 1.
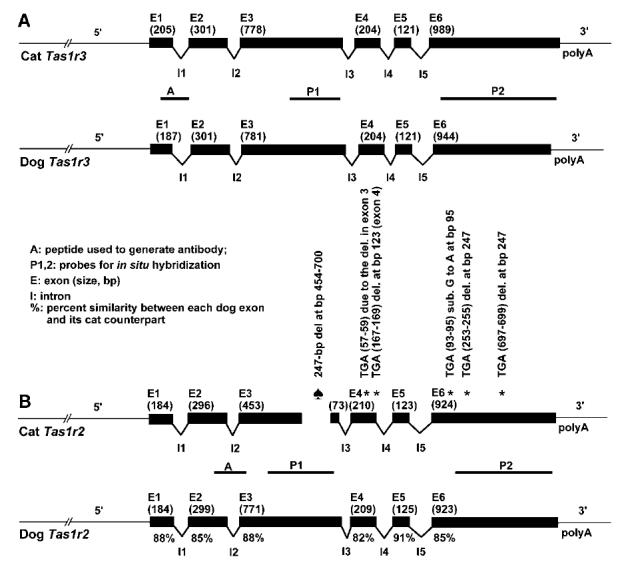
Gene structures of cat Tas1r3 and dog Tas1r3 (A), and cat Tas1r2 and dog Tas1r2 (B). The exons are shown in black (size in bp). Location (bp) refers to the position within each exon. Intron sizes shown in the figure are not proportionally scaled in (A) or (B) because of the large size of the Tas1r2 introns. Under each dog exon is the percentage of similarity between that exon and its cat counterpart at the nucleotide level (B). The exons for cat Tas1r2 refer to parts corresponding to dog exons. Asterisks indicate the position of microdeletion in exon 3 as well as the stop codon positions in exons 4 and 6 of cat Tas1r2. The accession for dog Tas1r3 is AY916759, and for dog Tas1r2 is AY916758.
We defined the exon-intron boundaries of cat Tas1r2 by comparison with known Tas1r2 from other species, e.g., humans and dogs. Within the sequence of cat Tas1r2, we discovered a microdeletion of 247 bp in exon 3. This deletion is responsible for a frame shift that results in a premature stop codon at bp 57-59 of exon 4 (Fig. 1b). By aligning cat Tas1r2 DNA sequences of exons 4, 5, and 6 with their dog counterparts, we found 4 additional stop codons: 1 in exon 4 due to a deletion at bp 123, and 3 in exon 6 due to a substitution at bp 95 and a deletion at bp 247 (Fig. 1b). The multiple stop codons indicate that the cat Tas1r2 is a pseudogene. In spite of using numerous (>70) primers corresponding to the message deduced from the Tas1r2 gene, we were unable to detect message of cat Tas1r2 from circumvallate and fungiform taste papillae.
RNA and protein expression
Having detected message from cat Tas1r3 but not from cat Tas1r2 by RT-PCR, we used the more tissue-specific approaches of in situ hybridization and immunohistochemistry to refine the search for cat Tas1r2 gene expression. The cat Tas1r3 gene was used as a positive control. The expression of Tas1r3 but not Tas1r2 in cat circumvallate papillae was confirmed by high-stringency in situ hybridization (15). To test for the presence of protein, cat circumvallate and fungiform papillae were exposed to polyclonal antibodies against T1R3 and T1R2. T1R3 labeling was present in the taste buds of circumvallate and fungiform papillae, whereas no T1R2 labeling was detected (15). These results suggest that Tas1r2 is not transcribed, or, if it is, it degrades rapidly, perhaps through a nonsense-mediated mRNA decay pathway (16), preventing synthesis of T1R2 protein.
Confirmation of Tas1r2 sequence in six individual cats, tiger and cheetah
We confirmed the sequence of Tas1r2 in 6 additional unrelated healthy adult domestic cats. Genomic DNA was amplified by PCR using primers that flanked the deletion and stop codons of the known cat Tas1r2, and sequenced. In addition, we performed PCR on genomic DNA of 1 tiger (Therion International) and 1 cheetah (a gift from the San Diego Zoo). We found that Tas1r2 in all 6 cats, the tiger, and the cheetah had the identical 247-bp deletion in exon 3, and had stop codons at the same positions in exon 4. In exon 6, we found evidence for 2 alleles at position 93-95 in domestic cats, wherein 2 cats show the stop codon, TGA (homozygotes TGA/TGA); 1 cat was heterozygote TGA/TGG; and 3 of the domestic cats, the tiger and the cheetah were homozygotes TGG/TGG. The second exon 6 stop codon is also common to all 3 species (TGA for domestic cats, TAG for tigers and cheetahs). Although the third stop codon of exon 6 at bp 697-699 occurred in all 6 domestic cats, the corresponding region in tigers and cheetahs could not be amplified by PCR.
These data are consistent with the supposition that cat Tas1r3 is an expressed and likely functional receptor, whereas cat Tas1r2 is an unexpressed pseudogene.
Sweet taste of cats and dogs
Earlier studies on sweet taste in cats and dogs reported that in contrast to cats, dogs prefer natural sugars, e.g., sucrose, glucose, fructose, and lactose, but not maltose (17-19). Dogs also show a preference for sodium cyclamate, but not for sodium saccharin (17,20). These comparative behavioral data are consistent with data generated from electrophysiological studies. Boudreau classified part of the cat taste system into several group units (I, II, IIIA, and IIIB). These cat units have their counterparts in the taste system of dogs (class B, A, C, and D units). Unlike cat group II units, dog class A units respond to sucrose and fructose (5). By recording from the chorda tympani nerve, Beidler found that cats do not respond to 0.5 mol/L sucrose, whereas dogs do (11). Anderson et al. (21) showed that taste nerve fibers responding to strychnine in dogs also respond to saccharin, which implies that dogs find saccharin aversive. Overall, cats and dogs respond very differently to sweet-tasting stimuli, although both species belong to Order Carnivora.
Taste and food selection
Taste receptors reflect a species’ food choices, and the genes encoding these receptors often show individual variation. These variations may or may not affect taste preference. A textbook example is the individual variation seen in sensitivity to the bitter compound, phenylthiocarbamide (PTC). A gene of the human TAS2R family of bitter taste receptors, TAS2R38, associated with this individual variation, shows 3 coding single-nucleotide polymorphisms giving rise to 5 haplotypes world-wide, accounting for the 55-85% of the variance in PTC sensitivity (22). In mice, variation in preference for sweet-tasting stimuli maps to the gene for T1R3, located within the Sac locus (23,24). This gene is allelic in mice, and several reports identify a missense mutation (I60T) as being the most likely mutation accounting for the phenotypic differences (12-14,25). However, the same alleles are not involved in strain-dependent sweet taste preference in rats (26).
In addition to the modulation of behavior that can be caused by point mutations, more profound behavioral changes can result from the abolishment of gene function through, for example, the generation of pseudogenes. An example of this effect in mammalian chemoreception lies within the large repertoire of olfactory receptor genes. Of the human olfactory receptor genes, >60% are pseudogenes (27), whereas only 20% are classified as such in mice (27,28). Strikingly, the accumulation of these olfactory pseudogenes in primates reportedly occurred concomitant with the acquisition of trichromatic color vision, perhaps reflecting the overarching behavioral changes that such an acquisition engendered (29). Similar generation of bitter taste receptor pseudogenes, accompanied by a large number of coding region single nucleotide polymorphisms, can account for the broad diversity displayed by the bitter taste receptor family. This diversity may play a role in both species-specific and individually manifested taste preference (30).
In the extreme case, in which a species fails to respond to stimuli representative of an entire modality, such as cats with sweet taste, the development of a unique food preference behavior, based on the remaining taste receptors, might be anticipated. With the exception of the sweetness modality, the taste system of the cat is organized much like that of most other mammals; thus, discovering the molecular basis for the lack of response to sweet tasting compounds in cats provides a window on the development of strict carnivorous behavior in Felidae.
ACKNOWLEDGMENTS
We thank Kirsten J. Mascioli and Minliang Zhou for technical assistance. We acknowledge Dr. Mark Haskins of the School of Veterinary Medicine, University of Pennsylvania and his laboratory for the procurement of animal tissue and the dedicated assistance of Patty O’Donnell and Karyn Cullen of that laboratory (NIH grants RR002512 and DK025759 to M.H.).
Footnotes
Published in a supplement to The Journal of Nutrition. Presented as part of The WALTHAM International Nutritional Sciences Symposium: Innovations in Companion Animal Nutrition held in Washington, DC, September 15-18, 2005. This conference was supported by The WALTHAM Centre for Pet Nutrition and organized in collaboration with the University of California, Davis, and Cornell University. This publication was supported by The WALTHAM Centre for Pet Nutrition. Guest editors for this symposium were D’Ann Finley, Francis A. Kallfelz, James G. Morris, and Quinton R. Rogers. Guest editor disclosure: expenses for the editors to travel to the symposium and honoraria were paid by The WALTHAM Centre for Pet Nutrition.
Author disclosure: V.L.D. is an employee of the Masterfoods division of Mars. G.K.B. is on an advisory board to the WALTHAM Centre. Patents describing the uses of the feline receptors are pending, and name as inventors: X.L., W.L., J.G.B., D.R.R., and A.A.B. Patents describing the uses of the canine receptors are pending, and name as inventors: X.L., W.L., and J.G.B.
Supported in part by The WALTHAM Centre for Pet Nutrition (to X.L. and J.G.B.), and by NIH grants R01DC00882 (G.K.B) and R03DC05154 (L.H.), and a grant from the National Science Foundation (DBJ-0216310 to N. Rawson). This project was also supported by a grant from the Pennsylvania Department of Health. The Department specifically disclaims responsibility for any analyses, interpretations, or conclusions. The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LITERATURE CITED
- 1.Beauchamp GK, Maller O, Rogers JG. Flavor preferences in cats (Felis catus and Panthera sp.) J Comp Physiol Psychol. 1977;91:1118–27. [Google Scholar]
- 2.Bartoshuk LM, Jacobs HL, Nichols TL, Hoff LA, Ryckman JJ. Taste rejection of nonnutritive sweeteners in cats. J Comp Physiol Psychol. 1975;89:971–5. doi: 10.1037/h0077172. [DOI] [PubMed] [Google Scholar]
- 3.Carpenter JA. Species differences in taste preference. J Comp Physiol Psychol. 1956;49:139–44. doi: 10.1037/h0048407. [DOI] [PubMed] [Google Scholar]
- 4.White TD, Boudreau JC. Taste preferences of the cat for neurophysiologically active compounds. Physiol Psychol. 1975;3:405–10. [Google Scholar]
- 5.Boudreau J, White T. Flavor chemistry of carnivore taste system. In: Society AC, editor. Flavor chemistry of animal foods. Washington, DC: 1978. pp. 102–28. [Google Scholar]
- 6.Boudreau JC, Bradley BE, Bierer PR, Kruger S, Tsuchitani C. Single unit recordings from the geniculate ganglion of the facial nerve of the cat. Exp Brain Res. 1971;13:461–88. doi: 10.1007/BF00234278. [DOI] [PubMed] [Google Scholar]
- 7.Boudreau JC, Oravec J, White TD, Madigan C, Chu SP. Geniculate neuralgia and facial nerve sensory systems. Arch Otolaryngol. 1977;103:473–81. doi: 10.1001/archotol.1977.00780250067007. [DOI] [PubMed] [Google Scholar]
- 8.Boudreau JC, Alev N. Classification of chemoresponsive tongue units of the cat geniculated ganglion. Brain Res. 1973;17:157–75. doi: 10.1016/0006-8993(73)90042-5. [DOI] [PubMed] [Google Scholar]
- 9.Dinger B, Fidone SJ, Stensaas FJ. Gustatory trophic action of arterial chemosensory neurones in the cat. J Physiol. 1984;356:49–64. doi: 10.1113/jphysiol.1984.sp015452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robinson PP. The characteristics and regional distribution of afferent fibres in the chorda tympani of the cat. J Physiol. 1988;406:345–57. doi: 10.1113/jphysiol.1988.sp017384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beidler LM, Fishman IY, Hardiman CW. Species differences in taste responses. Am J Physiol. 1955;181:235–9. doi: 10.1152/ajplegacy.1955.181.2.235. [DOI] [PubMed] [Google Scholar]
- 12.Bachmanov AA, Li X, Reed DR, Ohmen JD, Li S, Chen Z, Tordoff MG, de Jong PJ, Wu C, et al. Positional cloning of the mouse saccharin preference (Sac) locus. Chem Senses. 2001;26:925–33. doi: 10.1093/chemse/26.7.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reed DR, Li S, Li X, Huang L, Tordoff MG, Starling-Roney R, Taniguchi K, West DB, Ohmen JD, et al. Polymorphisms in the taste receptor gene (Tas1r3) region are associated with saccharin preference in 30 mouse strains. J Neurosci. 2004;24:938–46. doi: 10.1523/JNEUROSCI.1374-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Max M, Shanker YG, Huang L, Rong M, Liu Z, Campagne F, Weinstein H, Damak S, Margolskee RF. Tas1r3, encoding a new candidate taste receptor, is allelic to the sweet responsiveness locus Sac. Nat Genet. 2001;28:58–63. doi: 10.1038/ng0501-58. [DOI] [PubMed] [Google Scholar]
- 15.Li X, Li W, Wang H, Cao J, Maehashi K, Huang L, Bachmanov AA, Reed DR, Legrand-Defretin V, et al. Pseudogenization of a sweet-receptor gene accounts for cats’ indifference toward sugar. PLoS Genet. 2005;1:27–35. doi: 10.1371/journal.pgen.0010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rajavel KS, Neufeld EF. Nonsense-mediated decay of human HEXA mRNA. Mol Cell Biol. 2001;21:5512–9. doi: 10.1128/MCB.21.16.5512-5519.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferrell F. Preference for sugars and nonnutritive sweeteners in young beagles. Neurosci Biobehav Rev. 1984;8:199–203. doi: 10.1016/0149-7634(84)90041-1. [DOI] [PubMed] [Google Scholar]
- 18.Grace J, Russek M. The influence of previous experience on the taste behavior of dogs toward sucrose and saccharin. Physiol Behav. 1968;4:553–8. [Google Scholar]
- 19.Houpt KA, Coren B, Hintz HF, Hilderbrant JE. Effect of sex and reproductive status on sucrose preference, food intake, and body weight of dogs. J Am Vet Med Assoc. 1979;174:1083–5. [PubMed] [Google Scholar]
- 20.Houpt KA, Smith SL. Taste preferences and their relation to obesity in dogs and cats. Can Vet J. 1981;22:77–85. [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson B, Landgren A, Olsson L, Zotterman Y. The sweet taste fibers of the dog. Acta Physiol Scand. 1950;21:105–19. doi: 10.1111/j.1748-1716.1950.tb00718.x. [DOI] [PubMed] [Google Scholar]
- 22.Kim U, Jorgenson ECH, Leppert M, Risch N, Drayna D. Positional cloning of the human quantitative trait locus underlying taste sensitivity to phenylthiocarbamide. Science. 2003;299:1221–5. doi: 10.1126/science.1080190. [DOI] [PubMed] [Google Scholar]
- 23.Li X, Inoue M, Reed DR, Huque T, Puchalski RB, Tordoff MG, Ninomiya Y, Beauchamp GK, Bachmanov AA. High-resolution genetic mapping of the saccharin preference locus (Sac) and the putative sweet taste receptor (T1R1) gene (Gpr70) to mouse distal Chromosome 4. Mamm Genome. 2001;12:13–6. doi: 10.1007/s003350010236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X, Bachmanov AA, Li S, Chen Z, Tordoff MG, Beauchamp GK, de Jong PJ, Wu C, Chen L, et al. Genetic, physical and comparative map of the subtelomeric region of mouse chromosome 4. Mamm Genome. 2002;13:5–19. doi: 10.1007/s0033501-2109-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson G, Hoon MA, Chandrashekar J, Zhang Y, Ryba NJ, Zuker CS. Mammalian sweet taste receptors. Cell. 2001;106:381–90. doi: 10.1016/s0092-8674(01)00451-2. [DOI] [PubMed] [Google Scholar]
- 26.Lu K, McDaniel AH, Tordoff MG, Li X, Beauchamp GK, Bachmanov AA, VanderWeele DA, Chapman CD, Dess NK, et al. No relationship between sequence variation in protein coding regions of the Tas1r3 gene and saccharin preference in rats. Chem Senses. 2005;30:231–40. doi: 10.1093/chemse/bji019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilad Y, Man O, Paabo S, Lancet D. Human specific loss of olfactory receptor genes. Proc Natl Acad Sci U S A. 2003;100:3324–7. doi: 10.1073/pnas.0535697100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Young JM, Friedman C, Williams EM, Ross JA, Tonnes-Priddy L, Trask BJ. Different evolutionary processes shaped the mouse and human olfactory receptor gene families. Hum Mol Genet. 2002;11:535–46. doi: 10.1093/hmg/11.5.535. [DOI] [PubMed] [Google Scholar]
- 29.Gilad Y, Wiebe V, Przeworski M, Lancet D, Paabo S. Loss of olfactory receptor genes coincides with the acquisition of full trichromatic vision in primates. PLoS Biol. 2004;2:E5. doi: 10.1371/journal.pbio.0020005. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parry CM, Erkner A, le Coutre J. Divergence of T2R chemosensory receptor families in humans, bonobos, and chimpanzees. Proc Natl Acad Sci U S A. 2004;101:14830–4. doi: 10.1073/pnas.0404894101. [DOI] [PMC free article] [PubMed] [Google Scholar]


