Abstract
The active component of the licensed human anthrax vaccine (BioThrax™, or AVA) is a Bacillus anthracis toxin known as protective antigen (PA). Second generation anthrax vaccines currently under development are also based on a recombinant form of PA. Since the current and future anthrax vaccines are based on this toxin, it is important that the immunobiology of this protein in vaccinated humans be understood in detail. We have isolated and analyzed the PA-specific antibody repertoire from an AVA-vaccinated individual. When examined at the clonal level, we find an antibody response that is complex in terms of the combinatorial elements and immunoglobulin variable genes employed. All PA-specific antibodies had undergone somatic hypermutation and class switch recombination, both signs of affinity maturation. Although the antigenic epitopes recognized by the response were distributed throughout the PA monomer, the majority of antibodies arising in this individual following vaccination recognize determinants located on the amino-terminal (PA20) sub-domain of the molecule. This latter finding may have implications for the rational design of future PA-based anthrax vaccines.
Keywords: Bacillus anthracis, protective antigen, PA, antibody repertoire, repertoire analysis, anthrax, vaccine, human immune response, human monoclonal antibody
Introduction
The currently licensed anthrax vaccine (AVA or BioThrax™; Bioport Corporation, Lansing Michigan) consists of a sterile, bacteria-free filtrate prepared from a culture of a non-encapsulated Bacillus anthracis strain designated V770-NP1-R. In addition to various bacterial products, the vaccine is formulated to contain aluminum hydroxide as an adjuvant, benzethonium chloride as a preservative, and formaldehyde as a stabilizer (AVA, 2002). The primary immunogenic ingredient is the cell surface recognition component of the tripartite anthrax toxin complex known as protective antigen (PA). The vaccination series consist of three subcutaneous injections at 0, 2, and 4 weeks, and three booster vaccinations at 6, 12, and 18 months. Annual booster immunizations are recommended (AVA, 2002).
Although the vaccine itself is poorly characterized, a substantial body of evidence demonstrates that the toxin component PA is both necessary and sufficient to produce a protective antibody response following vaccination (Leppla et al., 2002). The undefined nature of AVA, along with the extended dosing schedule and requirement for yearly boosters have driven attempts to develop a more practical vaccine that is better characterized, well tolerated, and immunogenic. A vaccine containing purified recombinant PA (rPA) is currently under development as a replacement for AVA, and is in clinical trials to determine safety and immunogenicity. Since both AVA and the next generation vaccine are based on PA, it is important that the immunobiology of the human response to PA be understood in detail.
Reported here is the isolation and molecular analysis of the PA-specific antibody repertoire derived from an AVA-vaccinated individual who was enrolled in a CDC-sponsored clinical trial designed to address changes in route of administration and immunization regimens. The antibody response in this recipient was complex in terms of variable (V) gene usage, the combinatorial elements utilized, and the specific PA epitopes recognized. All PA-specific antibodies had undergone somatic hypermutation (SHM) and class switch recombination (CSR), both signs of affinity maturation. We have also determined that the majority of individual antibodies arising in this individual following vaccination recognize antigenic epitopes located in the amino-terminal (PA20) sub-domain of the PA monomer. This latter finding may have implications for toxin neutralization and the rational design of future PA-based anthrax vaccines.
Materials and Methods
Subjects
The donor analyzed in this report was recruited from individuals taking part in a larger study of the response to AVA being conducted at Baylor College of Medicine. Human subject protocols were reviewed and approved by the Institutional Review Boards at both Children's Hospital Oakland and Baylor College of Medicine.
Construction of Fab expression libraries
Fab expression libraries were constructed from MNCs enriched for PA-specific B cells in a manner similar to that previously described for polysaccharide-specific expression libraries (Reason et al., 1997; Reason and Zhou, 2006; Zhou et al., 2002; Zhou et al., 2004). PA, PA20, and PA63 were purchased from List Biological Laboratories, Campbell, CA. PA-specific Fabs were identified using a sensitive 125I-labeled PA capture assay and lysates of individual E. coli expression cultures. Positive isolates were re-cloned, heavy (H) and light (L) chain gene sequence determined, and PA-specific binding confirmed by ELISA. Initial sequence analysis utilized the NCBI IgBlast server (http://www.ncbi.nlm.nih.gov/igblast/) to identify candidate germline gene (Altschul et al., 1997). Subsequent analysis, alignments and translations were performed using MacVector (Accelrys Inc, Princeton, NJ). H and L chain V region gene nomenclature is as described in the IMGT database (Lefranc et al., 1999; Matsuda et al., 1998). Complementarity determining regions (CDRs) are as defined in (Kabat et al., 1991). Selected Fab clones were converted to full chain IgG1 antibodies and expressed in Chinese Hamster Ovary (CHO) cells using an in house PCI-derived bicistronic eukaryotic expression vector. Antibody was concentrated from the cell culture supernatant for use in binding assays.
Construction of PA20- and D4-GFP fusion proteins
The amino-terminal (residues 1-191) and the domain 4 carboxy-terminal (residues 587 – 735) portion of the PA monomer were cloned using PCR and expressed fused to intact green fluorescent protein (GFP). Cloning primers for the amino-terminal fragment were ATATGAATTCTATGGAAGTTAAACAGGAGAACCG (5') and ATATGGAT CCTCCTTCTACCTCTAATGAATC (3'). Cloning primers for the domain 4 region were GCATTAGAATTCGCATCACCATCACCATCACATGAATATTTTAATAAGA GATAAACG (5') and CGTATATCTAGAAGGATCCCCTATCTCATAGCCTTTTTTAGAAAAGAT (3'). Fusion proteins were expressed in E. coli and purified by nickel-chelate chromatography.
Domain specificity of PA-specific antibodies
The domain specificity of individual PA-specific antibodies was determined using capture assays, western blots of proteolytic fragments of PA, and western blots of PA20- and D4-GFP fusion proteins. In capture assays, 96-well plates coated with light chain-specific antibody were used to capture individual PA-specific antibodies. Plates were then washed and incubated with radio-labeled PA83, PA63, PA20, or D4-GFP. Binding was detected using PhosphorImager detection plates (Molecular Dynamics, Sunnyvale, CA). For western blots of proteolytic PA fragments, 1 μg each of PA83, PA63, and PA20 were electrophoresed using 4-12% Bis-Tris polyacrylamide gels (NuPAGE, Carlsbad, CA), electrically transferred to PVDF membranes, and probed with individual PA-specific antibodies. Binding was visualized by means of an alkaline-phosphatase conjugated goat antibody specific for human kappa or lambda light chains followed by BCIP/NBT color development. Western blots of PA20- and D4-GFP fusion proteins were processed in a similar fashion.
Antigen binding and Fab concentration assays
Fab concentration was determined by a capture ELISA in which goat anti-human Fd (The Binding Site, Birmingham, UK) or goat anti-IgA (Sigma, St. Louis, MO) immobilized on a microtiter plate captures Fab which is then detected by alkaline-phosphatase labeled goat anti-human L chain (Biosource International, Camarillo, CA). This assay is standardized with a purified Fab standard whose concentration was calculated from UV absorbance at 280 nm. A binding in ELISA was determined for both Fabs and full-chain IgG1 antibodies on 96-well plates coated with 5 μg/ml PA83 and developed with alkaline-phosphatase conjugated goat antibody specific for human kappa or lambda light chains.
Results
Approximately 6400 individual Fab clones were screened for antigen-specific binding. Fifty-nine PA-specific Fabs were isolated, of which 35 were unique in VH sequence, VL sequence, or both (summarized in Table 1). These clonal isolates bound PA in an antigen-specific and concentration dependent manner (Figure 1). The 35 unique Fabs represented 11 independent VH gene rearrangements (utilizing 8 different VH genes) and 11 unique VL rearrangements (utilizing 10 different VL genes). All V regions were mutated as compared to their germline gene of origin. Both kappa and lambda light chains were utilized in the response. In concordance with serum antibody, IgG1 was the predominant heavy chain isotype, although IgG2, IgG3, and IgA isotypes were also isolated. For ease of reference, the isolates have been grouped into families based on their unique VH gene rearrangement (Table 1).
TABLE 1.
Characteristics of the unique PA-specific Fab fragments
| Clone | VL | %Identitya | JL | L3(aa)b | IGHV | %Identitya | Isotype | JH | H3b | Familyc |
|---|---|---|---|---|---|---|---|---|---|---|
| 11A11 | IGKV1-5 | 96.5 | Jκ2 | 8 | IGHV1-18 | 95.8 | IgG1 | JH4 | 18 | H1-18 |
| 4A12 | IGKV1D-39 | 95.6 | Jκ4 | 9 | IGHV1-46 | 91.7 | IgG3 | JH4 | 12 | H1-46 |
| 34E3 | IGLV1-40 | 95.0 | Jλ3 | 12 | IGHV3-15 | 94.1 | IgG1 | JH1 | 13 | H3-15 |
| 35F7 | IGLV1-40 | 97.6 | Jλ3 | 12 | IGHV3-15 | 98.5 | IgG1 | JH1 | 13 | |
| 40D10 | IGLV1-40 | 94.7 | Jλ3 | 12 | IGHV3-15 | 96.3 | IgG1 | JH1 | 13 | |
| 41A3 | IGLV1-40 | 96.1 | Jλ3 | 12 | IGHV3-15 | 97.0 | IgG1 | JH1 | 13 | |
| 41C5 | IGLV1-40 | 96.5 | Jλ3 | 12 | IGHV3-15 | 93.7 | IgG1 | JH1 | 13 | |
| 46D1 | IGLV1-40 | 97.3 | Jλ3 | 12 | IGHV3-15 | 94.1 | IgG1 | JH1 | 13 | |
| 49F1 | IGLV1-40 | 96.8 | Jλ3 | 12 | IGHV3-15 | 94.1 | IgG1 | JH1 | 13 | |
| 51D6 | IGLV1-40 | 97.3 | Jλ3 | 12 | IGHV3-15 | 93.7 | IgG1 | JH1 | 13 | |
| 52E7 | IGLV1-40 | 97.5 | Jλ3 | 12 | IGHV3-15 | 91.5 | IgG1 | JH1 | 13 | |
| 53H4 | IGLV1-40 | 98.8 | Jλ3 | 12 | IGHV3-15 | 94.1 | IgG1 | JH1 | 13 | |
| 54C7 | IGLV1-40 | 99.1 | Jλ3 | 12 | IGHV3-15 | 94.8 | IgG1 | JH1 | 13 | |
| 55C7 | IGLV1-40 | 97.6 | Jλ3 | 12 | IGHV3-15 | 92.6 | IgA1 | JH1 | 13 | |
| 56F6 | IGLV1-40 | 95.0 | Jλ3 | 12 | IGHV3-15 | 94.1 | IgG1 | JH1 | 19 | |
| 59H10 | IGLV1-40 | 97.6 | Jλ3 | 12 | IGHV3-15 | 94.5 | IgG1 | JH1 | 13 | |
| 65A8 | IGLV1-40 | 98.8 | Jλ3 | 12 | IGHV3-15 | 94.1 | IgG1 | JH1 | 13 | |
| 65G8 | IGLV1-40 | 96.5 | Jλ3 | 12 | IGHV3-15 | 94.5 | IgG1 | JH1 | 13 | |
| 65H8 | IGLV1-40 | 97.6 | Jλ3 | 12 | IGHV3-15 | 94.8 | IgG1 | JH1 | 13 | |
| 66A5 | IGLV1-40 | 94.7 | Jλ3 | 12 | IGHV3-15 | 94.8 | IgG1 | JH1 | 13 | |
| 66B2 | IGLV1-40 | 95.9 | Jλ3 | 12 | IGHV3-15 | 93.7 | IgG1 | JH1 | 13 | |
| 17D2 | IGKV1-9 | 98.4 | Jκ4 | 9 | IGHV3-30 | 95.1 | IgG1 | JH3 | 15 | H3-30a |
| 65A3 | IGLV3-1 | 99.7 | Jλ3 | 9 | IGHV3-30 | 94.3 | IgG1 | JH6 | 17 | H3-30b |
| 47F12 | IGLV1-44 | 96.7 | Jλ3 | 11 | IGHV3-30 | 94.0 | IgG1 | JH6 | 18 | H3-30c |
| 59G5 | IGLV1-44 | 97.0 | Jλ3 | 11 | IGHV3-30 | 94.0 | IgG1 | JH6 | 18 | |
| 8H2 | IGKV6-21 | 96.3 | Jκ2 | 9 | IGHV3-33 | 94.7 | IgG1 | JH4 | 13 | H3-33a |
| 19E1 | IGKV6-21 | 99.1 | Jκ2 | 9 | IGHV3-33 | 94.7 | IgG1 | JH4 | 13 | |
| 41F12 | IGLV1-47 | 97.3 | Jλ1 | 11 | IGHV3-33 | 95.1 | IgG1 | JH4 | 12 | H3-33b |
| 55F9 | IGLV1-47 | 97.0 | Jλ1 | 11 | IGHV3-33 | 95.8 | IgG1 | JH4 | 12 | |
| 20C7 | IGKV1-9 | 99.4 | Jκ2 | 9 | IGHV3-43 | 95.8 | IgG1 | JH6 | 17 | H3-43 |
| 51C5 | IGLV3-21 | 97.6 | Jλ2 | 11 | IGHV3-74 | 95.8 | IgG1 | JH4 | 14 | H3-74 |
| 1A5 | IGKV2-30 | 97.6 | Jκ1 | 9 | IGHV4-59 | 90.8 | IgG1 | JH1 | 11 | H4-59 |
| 1B10 | IGKV2-30 | 97.3 | Jκ1 | 9 | IGHV4-59 | 90.8 | IgA1 | JH1 | 11 | |
| 4B1 | IGKV2-30 | 97.3 | Jκ1 | 9 | IGHV4-59 | 90.8 | IgG1 | JH1 | 11 | |
| 13E12 | IGKV2-30 | 97.3 | Jκ1 | 9 | IGHV4-59 | 90.8 | IgG2 | JH1 | 11 |
Percent nucleotide identity over the entire V region as compared to the corresponding germline gene.
Number of residues comprising the VL and IGHV CDR3 region; aa = amino acids.
Family denotes unique IGHV rearrangements
Figure 1.
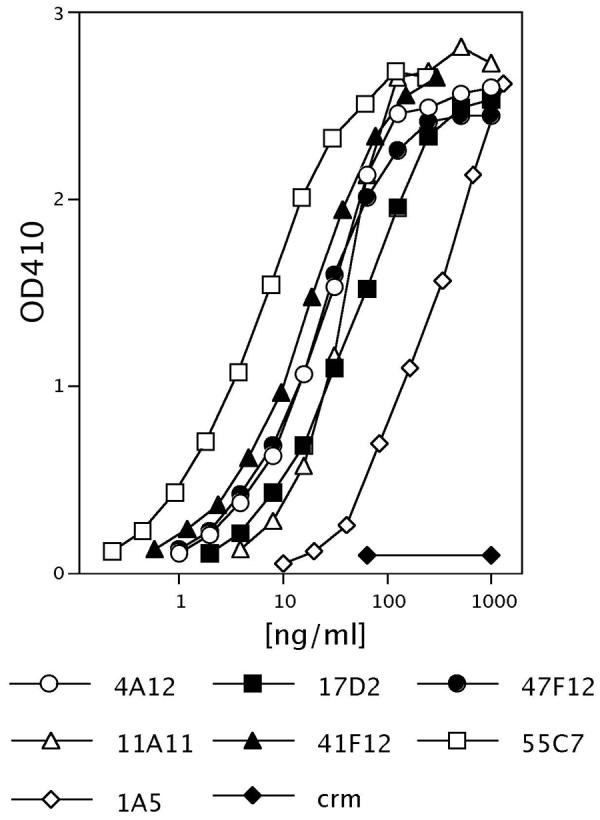
Protective antigen binding by representative PA-specific human monoclonal antibodies in an ELISA assay. All antibodies bound PA in a concentration dependent manner and did not bind the mutant diphtheria toxin CRM197 negative control toxin at any concentration tested.
Somatic hypermutation
All VH genes (Table 2) and VL genes (Table 3) were mutated as compared to their germline gene of origin. Mutations were seen in both the complementarity determining regions (CDRs) as well as the framework regions of the V genes. Mutations were rarely observed in the light or heavy chain constant region (average of 0.3 mutations/constant region), indicating that the observed mutations are most likely generated during somatic hypermutation and are unlikely to have arisen as PCR artifacts. The majority of VL mutations (52%) and 47% of VH mutations were associated with the RGYW/WRCY sequence motifs recognized as “hotspots” of mutation during activation-induced cytidine deaminase (AID)-mediated hypermutation of immunoglobulin genes (Wagner and Neuberger, 1996). These frequencies are roughly twice that of the occurrence of these motifs in the germline V genes. The isolation of several clonally related members of the IGHV3-15 family illustrates the degree to which the PA-specific antibody repertoire can be diversified by somatic hypermutation, even within the progeny of a single B cell rearrangement event. The VH members of this family had highly similar CDR3 regions, were all rearranged with JH1, and shared several mutations as compared with the IGHV3-15 germline gene. The VL members of this family were all rearranged with Jλ3, had a non-encoded residue inserted at the V/J junction, and shared several mutations when compared to the IGLV1-40 lambda germ line gene of origin. It is therefore likely that the members of this family derived from a single initial rearrangement event and diverged in sequence as a result of somatic hypermutation during clonal expansion. It should be noted that only those derivative clones retaining affinity for antigen (PA) would be detected using our methodology.
TABLE 2.
H chain CDRsa
| Clone | CDR1 | CDR2 | CDR3 | J | Isotype |
|---|---|---|---|---|---|
| IGHV1-18 | SYGIS | WISAYNGNTNYAQKLQG | |||
| 11A11 | NF--T | Y-------A-------- | DRGSRWFGEFPDEYYFDY | JH4 | IgG1 |
| IGHV1-46 | SYYMH | IINPSGGSTSYAQKFQG | |||
| 4A12 | S-FV- | M---R----N------- | VNWAYGDYDFDY | JH4 | IgG3 |
| IGHV3-15 | NAWMS | RIKSKTDGGTTDYAAPVKG | |||
| 35F7 | D---T | ---R--------------- | DVLGIVIIVGAAH | JH1 | IgG1 |
| 40D10 | K---- | ---R-S------------- | ------------- | JH1 | IgG1 |
| 59H10 | Q-H-- | -M-R-S------------- | --R---------- | JH1 | IgG1 |
| 46D1 | Q-H-- | -M-R-S-------G----- | --R---------- | JH1 | IgG1 |
| 41A3 | Y-S-T | ---R--------------- | --R---------- | JH1 | IgG1 |
| 49F1 | K---T | ---RRS------------- | --------R---Q | JH1 | IgG1 |
| 55C7 | Y-R-- | ---R--E------------ | ------------- | JH1 | IgG1 |
| 52E7 | Y-R-- | ---N-R--E--H------- | ------------- | JH1 | IgG1 |
| 66A5 | K---T | ---R-S----I-------- | --R---------- | JH1 | IgG1 |
| 34E3 | K---T | ---RRS------------- | --------R---Q | JH1 | IgG1 |
| 41C5 | ----T | ---RRS------------- | ------------Q | JH1 | IgG1 |
| 51D6 | ----T | ---RRS------------- | ------------Q | JH1 | IgG1 |
| 53H4 | Q-H-- | -M-R-S-------G----- | --R---------- | JH1 | IgG1 |
| 54C7 | ----- | ---R-S----I----S--- | ------------Q | JH1 | IgA1 |
| 56F6 | K---T | ---RRS------------- | --------R---Q | JH1 | IgG1 |
| 65A8 | ----T | ---RRS------------- | ------------Q | JH1 | IgG1 |
| 65H8 | ----- | ---R-S----I----S--- | ------------Q | JH1 | IgG1 |
| 65G8 | D---T | ---R-S------------- | -T----R------ | JH1 | IgG1 |
| 66B2 | ----T | ---RRS------------- | ------------Q | JH1 | IgG1 |
| IGHV 3-30 | SYGMH | VISYDGSNKYYADSVKG | |||
| 17D2 | R---- | ------GD-K-V----- | WDYVWESYRGKAFDI | JH3 | IgG1 |
| 65A3 | --A-- | L-----NT--------- | LISYDGNTKYYADSVKG | JH6 | IgG1 |
| 47F12 | --T-- | L-----T--N-G---T- | ARVIVPAGSNYNQYGMDV | JH6 | IgG1 |
| 59G5 | --T-- | L-----T--N-G---T- | ARVIVPAGSNYNQYGMDV | JH6 | IgG1 |
| IGHV 3-33 | SYGMH | VIWYDGSNKYYADSVKG | |||
| 8H2 | DF--- | L--N---K--------- | WGYYYGSGSPPEY | JH4 | IgG1 |
| 19E1 | DF--- | L--N---K--------- | WGYYYGSGSPPEY | JH4 | IgG1 |
| 41F12 | T---- | ---F------------- | EDGSYHQGPFDY | JH4 | IgG1 |
| 55F9 | T---- | ---F------------- | EDGSYHQGPFDY | JH4 | IgG1 |
| IGHV 3-43 | DYTMH | LISWDGGSTYYADSVKG | |||
| 20C7 | ----- | --T----R-F--N---- | GPNRRDSFGLHYYGLDV | JH6 | IgG1 |
| IGHV 3-74 | SYWMH | RINSDGSSTSYADSVKG | |||
| 51C5 | R-Y-- | R--N------------- | EGRPMGLGTSVGMN | JH4 | IgG1 |
| IGHV 4-59 | SYYWS | YIYYSGSTNYNPSLKS | |||
| 1A5 | -HF- | ---SG-T---D----- | GDMVTGDPGDY | JH1 | IgG1 |
| 1B10 | -HF- | ---SG-T---D----- | ----------- | JH1 | IgA1 |
| 4B1 | -HF- | ---SG-T---D----- | ----------H | JH1 | IgG1 |
| 13E12 | -HF- | ---SG-T---D----- | ----------- | JH1 | IgG2 |
Heavy chain CDR residues and gene usage of PA-specific Fabs. Translation of germ line CDRs shown for comparison.
TABLE 3.
L chain CDRsa
| Clone | CDR1 | CDR2 | CDR3 | J | |
|---|---|---|---|---|---|
| IGKV2-30 | RSSQSLVYSDGNTYLN | KVSNRDS | MQGTHWP | ||
| 1A5 | -------S---K---- | ---K--- | --A---L | WT | JK1 |
| IGKV6-21 | RASQSIGSSLH | YASQSFS | HQSSSLP | ||
| 8H2 | -------RN-- | -V----- | -----F- | YT | JK2 |
| 19E1 | --------N-- | ------- | -----F- | YT | JK2 |
| IGKV1-5 | RASQSISSWLA | DASSLES | QQYNSYS | ||
| 11A11 | ---H---T--- | -A---Q- | ---S-Y | YT | JK2 |
| IGKV1-9 | RASQGISSYLA | AASTLQS | QQLNSYP | ||
| 17D2 | -------R--- | ------G | ------- | LT | JK4 |
| 20C7 | ----E------ | ------- | ------- | YT | JK2 |
| IGKV1D-39 | RASQSISSYLN | AASSLQS | QQSYSTP | ||
| 4A12 | ------AR--- | D----HT | --T---- | LT | JK4 |
| IGLV1-40 | TGSSSNIGAGYDVH | GNSNRPS | QSYDSSLSG | ||
| 51D6 | ----------K--- | ------- | --------- D | WV | JL3 |
| 34E3 | ---F--V-S-S--- | --T---- | -----R--- D | WV | JL3 |
| 35F7 | ----------N--- | ------- | --------- D | WV | JL3 |
| 41A3 | ---G------S--- | ------- | ---G----- D | WV | JL3 |
| 65H8 | ----------N--- | ------- | --------- D | WV | JL3 |
| 65A8 | ----------S--- | ------- | --------- D | WV | JL3 |
| 52E7 | ----------N--- | ------- | --------- D | WV | JL3 |
| 53H4 | ----------S--- | ------- | --------- D | WV | JL3 |
| 54C7 | ----------S--- | ------- | ----R---- D | WV | JL3 |
| 46D1 | ----------S--- | ------- | -----R-G- E | WV | JL3 |
| 55C7 | ----------S--- | ------- | -----R-G- E | WV | JL3 |
| 59H10 | ----------S--- | ------- | -----R-G- E | WV | JL3 |
| 65G8 | ---G------S--- | ------- | ---G----- D | WV | JL3 |
| 41C5 | --D---M---S--- | ------- | --------- D | WV | JL3 |
| 56F6 | ---F--V-S-S--- | --T---- | -----R--- D | WV | JL3 |
| 40D10 | ---F--V-S-S--- | --T---- | -----R--- D | WV | JL3 |
| 66A5 | ---F--V-S-S--- | --T---- | -----R--- D | WV | JL3 |
| 49F1 | ----------S--- | --T---- | ----R---- D | WV | JL3 |
| 66B2 | ---L------K--- | ------- | ----N---- D | WV | JL3 |
| IGLV1-44 | SGSSSNIGSNTVN | SNNQRPS | AAWDDSLNG | ||
| 47F12 | ---W--T-R---- | --T---- | -------K- | WM | JL3 |
| 59G5 | ---W----R-A-- | T-T---- | -------K- | WV | JL3 |
| IGLV1-47 | SGSSSNIGSNYVY | SNNQRPS | AAWDDSLSG | ||
| 41F12 | ---------H--C | R-D---- | -S-----T- | QV | JL1 |
| 55F9 | ---------H--C | R-D---- | -S-----T- | QV | JL1 |
| IGLV3-1 | SGDKLGDKYAC | QDSKRPS | QAWDSSTA | ||
| 65A3 | ----------- | ------- | -------- | T | JL3 |
| IGLV3-21 | GGNNIGSKSVH | DDSDRPS | QVWDSSSDH | ||
| 51C5 | ------R---- | --R---- | ----NI--- | VI | JL2 |
Light chain CDR residues and gene usage of PA-specific Fabs. Translation of germ line CDRs shown for comparison.
Class Switch Recombination
The heavy chain isotype of serum antibody directed against PA is primarily IgG1. Serum antibodies directed against protein antigens are in general dominated by the IgG1 isotype (Hammarstrom et al., 1987; Papadea and Check, 1989). Thirty-one of the 35 paratopes reported herein are also IgG1 in concordance with this fact. A single member of the IGHV3-15 family (55C7; Table 1) had undergone secondary CSR to IgA1. The sharing by this clone of the CDR3 sequence and several mutations with the IgG1 isolates of this family (Table 2) indicates this secondary CSR occurred in a IgG1 precursor, and not directly from the (presumed) IgM progenitor during the course of ongoing SHM. The alternative interpretation, that of an IgA1 precursor giving rise to the IgG1 clonal isolates is less likely, due to the relative position of the IgG1 and IgA1 constant region genes in the germline, and the mechanism responsible for CSR which results in the deletion of intervening gene sequences (Jack et al., 1988; Mills et al., 1992). The lack of sibling VH sequences for the single IgG3 isolate (4A12; Table 1) prevents placement of this CSR in the context of ongoing SHM. The IGHV4-59 family sequences (Table 2) are notable in that, although all are mutated as compared to the germline gene of origin, the high degree of sequence similarity in the 4 members of this group indicate that CSR giving rise to IgG1, IgG2, and IgA1 isotypes occurred late in the divergence of these clones, and without additional SHM following class switch.
Sub-domain specificity
The PA molecule is an 83 kd protein and as such would be expected to present a variety of antigenic epitopes to the immune system. The domain structure of PA is understood in detail (Brossier et al., 2000; Little and Lowe, 1991; Petosa et al., 1997; Singh et al., 1991), and various functional aspects of the molecule have been ascribed to the various sub-domains of the molecule (Ahuja et al., 2001; Cunningham et al., 2002; Novak et al., 1992). Radio-labeled capture assays and western blots were utilized to determine the domain specificity of the various PA-specific paratopes described in this study. In addition to the purified proteolytic fragments PA63 and PA20, GFP-fusion proteins containing the amino-terminal PA20 domain (residues 1-191) and the caboxy-terminal domain 4 (residues 587-735) were constructed, expressed, and purified for use in capture assays and Western blots. The results of these assays are shown in Table 4. As would be expected, domain specificity was consistent within those paratope families in which multiple members were available for analysis. Seven of the 11 paratope families were specific for epitopes found in the PA20 portion of the molecule. Two bound to epitopes found in domain 4, and 2 bound to residues within PA63 but not associated with domain 4. Although the three assays used (capture assay, western blot of fusion proteins, western blot of proteolytic fragments) were not contradictory for any of the assayed paratopes, there were a few cases where they were not internally consistent. Clone 11A11, for example, bound PA20 in the capture assay, and the PA20-GFP fusion protein in the Western blot, but did not bind the PA20 proteolytic fragment in the western blot. We believe these cases are indicative of an alteration of epitope structure in the different methods of fragment preparation and presentation, and highlight the necessity of utilizing multiple methodologies when assaying epitope specificity.
Table 4.
PA Domain Binding by PA-specific Fabs
| Capture assay |
Western blots |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Clone | VL | VH | PA83a | PA63a | PA20a | fD4b | fD1c | fD4b | PrFraga |
| 11A11 | IGKV1-5 | IGHV1-18 | + | − | + | nd | + | − | - |
| 4A12 | IGKV1D-39 | IGHV1-46 | + | + | − | + | − | + | PA63 |
| 35F7 | IGLV1-40 | IGHV3-15 | + | − | +/− | − | + | − | PA20 |
| 40D10 | IGLV1-40 | IGHV3-15 | + | − | + | − | + | − | PA20 |
| 59H10 | IGLV1-40 | IGHV3-15 | + | − | + | − | + | − | PA20 |
| 46D1 | IGLV1-40 | IGHV3-15 | + | − | + | − | + | − | PA20 |
| 41A3 | IGLV1-40 | IGHV3-15 | + | − | +/− | − | + | − | PA20 |
| 49F1 | IGLV1-40 | IGHV3-15 | + | − | − | − | +/− | − | - |
| 55C7 | IGLV1-40 | IGHV3-15 | + | − | + | − | + | − | nd |
| 52E7 | IGLV1-40 | IGHV3-15 | + | − | + | − | + | − | nd |
| 66A5 | IGLV1-40 | IGHV3-15 | + | − | + | − | + | − | PA20 |
| 34E3 | IGLV1-40 | IGHV3-15 | + | − | + | − | + | − | PA20 |
| 41C5 | IGLV1-40 | IGHV3-15 | + | − | +/− | − | + | − | PA20 |
| 51D6 | IGLV1-40 | IGHV3-15 | + | − | + | − | + | − | PA20 |
| 53H4 | IGLV1-40 | IGHV3-15 | + | − | + | − | + | − | PA20 |
| 54C7 | IGLV1-40 | IGHV3-15 | + | − | + | − | + | − | nd |
| 56F6 | IGLV1-40 | IGHV3-15 | + | − | + | − | + | − | PA20 |
| 65A8 | IGLV1-40 | IGHV3-15 | + | − | + | − | + | − | nd |
| 65H8 | IGLV1-40 | IGHV3-15 | + | − | + | nd | + | − | PA20 |
| 65G8 | IGLV1-40 | IGHV3-15 | + | − | + | − | + | − | PA20 |
| 66B2 | IGLV1-40 | IGHV3-15 | + | − | + | − | + | − | nd |
| 17D2 | IGKV1-9 | IGHV3-30 | + | + | − | − | − | − | PA63 |
| 65A3 | IGLV3-1 | IGHV3-30 | + | − | +/− | − | + | − | nd |
| 47F12 | IGLV1-44 | IGHV3-30 | + | − | + | − | + | − | PA20 |
| 59G5 | IGLV1-44 | IGHV3-30 | + | − | + | − | + | − | PA20 |
| 8H2 | IGKV6-21 | IGHV3-33 | + | − | + | − | + | − | PA20 |
| 19E1 | IGKV6-21 | IGHV3-33 | + | − | + | nd | + | − | PA20 |
| 41F12 | IGLV1-47 | IGHV3-33 | + | − | + | − | + | − | PA20 |
| 55F9 | IGLV1-47 | IGHV3-33 | + | − | + | − | + | − | PA20 |
| 20C7 | IGKV1-9 | IGHV3-43 | + | + | − | + | − | + | - |
| 51C5 | IGLV3-21 | IGHV3-74 | + | − | + | − | − | − | - |
| 1A5 | IGKV2-30 | IGHV4-59 | + | + | − | − | − | − | PA63 |
| 1B10 | IGKV2-30 | IGHV4-59 | + | +/− | − | − | − | − | PA63 |
| 4B1 | IGKV2-30 | IGHV4-59 | + | − | − | − | − | − | nd |
| 13E12 | IGKV2-30 | IGHV4-59 | + | +/− | − | − | − | − | nd |
Purified PA83 or purified PA63 or PA20 proteolytic fragments.
Purified Domain 4-GFP fusion protein.
Purified Domain 1(PA20)-GFP fusion protein.
Discussion
The currently approved vaccine used for the prevention of anthrax infection is safe and, to the degree to which it can be tested, effective (Friedlander et al., 1999; Sever et al., 2004; Sever et al., 2002). It has several shortcomings, however. The requirement of an extended 18 month immunization regimen before full protection is achieved, the necessity of yearly booster vaccinations to maintain protective levels of serum antibody, and the perception (although unsupported) of adverse side effects associated with immunization have driven attempts to develop a new, or “second generation” vaccine formulation with improved performance characteristics (Brey, 2005; Leppla et al., 2002). These vaccines have recombinant PA as their active component, and are currently in clinical trials. Initial results suggest little improvement in immunogenicity as compared to AVA (Gorse et al., 2006). An understanding of the immunobiology of the human antibody response to PA at the molecular level may facilitate the more rational design of a vaccine candidate.
The biology of the anthrax toxin system has been extensively studied and is understood in detail (reviewed in (Brossier and Mock, 2001; Leppla, 1995)). PA is an 83 kd bi-functional protein that recognizes the widely distributed cell surface receptors TEM8 and CCMG and provides cell targeting (Bradley et al., 2001; Scobie et al., 2003). Binding is rapidly followed by furin-mediated cleavage of PA83 at the RKKR motif (residues 164-167) to produce an amino terminal fragment (PA20), and the remainder of the PA monomer (PA63) (Brossier and Mock, 2001). PA20 is thought to have no further role in intoxication. Cleavage allows PA63 to form ring-shaped heptamers on the cell surface, and also exposes binding sites for the other two members of the B. anthracis toxin system, Lethal Factor (LF) and Edema factor (EF). Following heptamerization and LF and/or EF binding, the entire complex is internalized by means of receptor mediated endocytosis (Brossier and Mock, 2001). Once within the acidic environment of the endocytic vacuole, the lytic components of the toxin system are actively transported across the endocytic vacuole membrane into the cytoplasm of the cells (Ren et al., 2004). Lethal factor (776 residues) is a zinc protease with specificity for several members of the mitogen-activated protein kinase (MAPKK) family (Brossier and Mock, 2001). Edema factor (767 residues) is an adenylate cyclase and provokes a substantial increase in intracellular cAMP (Brossier and Mock, 2001).
Our research utilizes repertoire cloning as a methodology for analyzing antigen-specific antibody response in humans (Lucas et al., 2001; Reason and Zhou, 2004; Zhou et al., 2002; Zhou et al., 2004). In the case of complex immunogens such as proteins, it allows us to analyze the monoclonal components of the antibody response in terms of their individual epitopes and functionality. By analyzing different individuals in a population, we can also determine the degree to which different members of the population utilize the same mechanisms in generating antibody diversity. A shortcoming of this approach is that native H and L chain pairing is lost. We have shown through direct protein sequencing and idiotypic analysis that repertoire cloning faithfully reproduces the serum repertoire of polysaccharide-specific antibodies that arise in vaccinated individuals (Reason et al., 1997; Zhou et al., 2002; Zhou et al., 2004), and that in these systems de novo antigen-specific paratopes are not created by this methodology. Protein-specific antibody repertoires are more diverse and, as demonstrated in this study, even a highly enriched protein-specific B cell population contains multiple paratopes utilizing several different H and L chains. We cannot discount that combinatorial pairing in such a complex mixture might give rise to an antigen-binding Fab not present in the in vivo population, especially in cases where the majority of the contact residues are located on one chain. We have therefore eliminated from analysis those Fabs in which V gene pairing deviated from the majority of isolates for that family and which demonstrated reduced binding affinity for PA. This exclusion criteria is informal, however, and the data must be interpreted with this in mind. And, although our analysis was fairly extensive, it is likely that screening of additional clones from the expression library would reveal additional PA-specific Fabs. As a result of these two facts, we may have underrepresented the complexity of the response in this donor.
Combinatorial Diversity in the response to PA
In addition to the recruitment of 8 VH and 10 VL germline genes, a variety of previously understood mechanisms combined to diversify the initial response to PA in this individual. Three different rearrangements of the IGHV3-30 germline gene with two J region gene segments (Table 2) generate substantial diversity in the H chain CDR3 region, both in terms of amino acid sequence and in length. Likewise, two separate IGHV 3-33 rearrangements were identified based on their differing CDR3 sequences and lengths. The three IGHV3-30 rearrangements pair with different VL genes (both kappa and lambda) as do the two IGHV3-33 rearrangements. The insertion of non-encoded residues at the V-J junction (the IGLV1-40 lambda L chains), as well as the deletion of germline encoded residues at this junction (the IGLV3-1 lambda L chain), have been demonstrated in other antigen-specific repertoires and are examples of diversifying events that take place during the initial stages of V gene rearrangement. VJ and VDJ joining, VH/VL pairing, VH CDR3 generation, and the insertion and/or deletion of residues at the V/J junction are all examples of diversification that occur independent of antigenic stimulation early in B cell development.
Somatic hypermutation in the response to PA
A major source of diversity in this donor's overall response to PA was somatic hypermutation. SHM is antigen driven and therefore occurs following vaccination or infection. SHM is also believed to require the recruitment of T cells or T cell derived cytokines (Miller et al., 1995). All of the V genes utilized by these paratopes were mutated as compared to their germline gene of origin. Mutations were restricted to the V regions in both VH and VL sequences, and were highly correlated with the RGYW/WRCY motifs that are the established targets of AID-mediated SHM (Wagner and Neuberger, 1996). Mutations were not confined to the CDRs and occurred in the framework regions as well. It is difficult to recognize mutations that occur in the CDR3 region of the VH gene, as there are usually no well-defined germline elements available for comparison. Our isolation of several members of the IGHV3-15 family allow us to demonstrate that SHM occurs in the VH CDR3 region as well. The lack of mutations in the L chain constant region and the CH1 region of the H chains indicates that these mutations are physiologic in origin, and not the result of PCR artifact. In the two cases where multiple rearrangements of the same germline genes were isolated (the IGHV 3-30 and IGHV 3-33 families), there was a tendency for mutations to target the same CDR residues across the different rearrangements (Table 2), possibly indicating residues that are in contact with the antigen.
Class switch recombination
IgG1 is the principal antigen-specific antibody isotype in hyperimmune antisera raised to protein antigens such as PA. This isotype bias is reflected in the predominance of IgG1 in the Fabs we isolated from this individual. It is of interest to note that in the IGHV4-59 family where secondary CSR did occur (Table 1), SHM was apparently restricted to that period prior to the class switch, resulting in antibodies with mutated but identical paratopes associated with IgG1, IgG2, and IgA1 isotypes. All IGKV2-30 light chain members of this family were likewise identical in sequence. It is likely that SHM in these clones terminated at the point of secondary CSR. Although more difficult to interpret, the single IgA1 member of the IGHV3-15 family (Table 2) is identical in V region sequence to the IgG1 clone 65H8, and may represent another example of SHM terminating following secondary CSR. This is in contrast to what has been reported for the response to carbohydrate antigens where CSR has been shown to occur throughout the course of ongoing SHM (Reason and Zhou, 2006; Zhou et al., 2002; Zhou et al., 2004).
Domain specificity of the component paratopes
The serum antibody response to any complex protein antigen is the summation of contributions made by the individual responding B cell clones. The analytical techniques we employ allow us to characterize both the structure and the contribution made by each of the individual paratopes to the total antibody response. We used several different assays to access the domain specificity of the paratopes we isolated. Since all assays involve some modification or possible partial denaturation of the antigen, the use of multiple assays increases the likelihood of preserving targeted epitopes. In addition, the PA20-GFP fusion protein was designed to retain the furin cleavage site, thereby allowing us to detect epitopes that span that junction. Together, these techniques allowed us to unambiguously assign each paratope to a corresponding region of the toxin. Although low in resolution, epitope mapping at the domain level permits an estimation of the minimum number of epitopes recognized in the response, and suggest mechanisms by which they might function to neutralize toxicity.
The procedures we employ to generate and identify PA-specific Fabs may in some way bias our results towards paratopes that bind a particular sub-domain of the molecule, but we believe this to be unlikely. The PA used for biotinylation, cell selection, radio-iodination, and screening is a highly purified 83 kd molecule that appears on silver-stained PAGE gels to be free of any significant contamination with smaller proteolytic fragments. Residues available for biotinylation and radio-iodination are evenly distributed throughout the molecule. Individual colonies are screened by capturing 125I-PA from solution to maximize the epitope integrity of the antigen. The complex distribution of H and L V genes utilized in the PA-specific response make it unlikely that an epitope bias could be introduced during the initial PCR reaction based on primer design. Taken together, these factors suggest that the epitope distribution we observe for our isolated Fabs reflects the epitope distribution present in the ongoing immune response in the vaccinated individual.
Paratopes were identified that recognized epitopes in domain 4, in PA63 (excluding domain 4), and in PA20. As would be expected, in those families with multiple diversified isolates, domain specificity was conserved across all isolates, although relative affinity appeared to decrease in some due to their accumulated somatic mutations. Although the PA20 domain of the molecule constitutes less than 25% of the mass of the intact monomer, a majority (7/11) of the paratope families isolated from this individual react exclusively with antigenic epitopes associated with this amino-terminal sub-domain. If this epitope distribution is true for the majority of vaccine recipients, it may have implications for vaccine effectiveness. PA20 is cleaved from the remaining part of the molecule immediately after binding to the cell surface receptor, and is not thought to play any further role in intoxication. Although antibodies binding to the PA20 region of the molecule could conceivably block furin mediated cleavage and thus be protective, it is also possible that although these antibodies bind PA with high avidity, they are non-functional in terms of toxin neutralization. The mechanism responsible for this bias in epitope distribution is unknown, and may include factors intrinsic to the primary amino acid sequence of the two fragments. However, it may also arise from differences in antigen processing and peptide presentation. PA20 most likely enters the antigen processing pathway through the route established for foreign proteins (Brodsky and Guagliardi, 1991). PA63, on the other hand, directs its own entry to the cell. It is possible that these two different entry pathways result in a difference in the efficiency of peptide presentation. The bias in antibody epitope specificity towards determinants associated with PA20 might therefore arise as a secondary result deriving from this difference in antigen presentation.
Conclusion
Our findings in this one individual indicate that the anthrax vaccine formulation currently in use recruits a diverse variety of responding B cell of clones.Furthermore, vaccination with AVA induces clonal expansion, somatic hypermutation, and class-switch recombination in antibodies specific for the immunogenic toxin component PA. These activities are consistent with the generation of a mature and efficacious antibody response. We also find the percentage of antibodies reactive with PA20-associated epitopes to be disproportionate to the size of this proteolytic fragment, raising the possibility that many of the individual antibody species that make up the serological response are not capable of neutralizing toxin function. Additional donors are currently being analyzed to determine if this epitope bias is characteristic of the PA-specific response in general.
Acknowledgments
The authors gratefully acknowledge Nanette Bond, PA-C for assistance with sample collection and Betty M. Ho for critically reading the manuscript.This work was supported by Public Health Service Grants AI57932 and AI066508 from the National Institute of Allergy and Infectious Diseases. This research was conducted in a facility constructed with support from Research Facilities Improvement Program Grant Number C06 RR-16226 from the National Center for Research Resources, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- AVA 2002. Anthrax Vaccine Adsorbed, Biothrax™. Package Insert.
- Ahuja N, Kumar P, Bhatnagar R. Hydrophobic residues Phe552, Phe554, Ile562, Leu566, and Ile574 are required for oligomerization of anthrax protective antigen. Biochem. Biophys. Res. Commun. 2001;287:542–549. doi: 10.1006/bbrc.2001.5613. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley KA, Mogridge J, Mourez M, Collier RJ, Young JA. Identification of the cellular receptor for anthrax toxin. Nature. 2001;414:225–229. doi: 10.1038/n35101999. [DOI] [PubMed] [Google Scholar]
- Brey RN. Molecular basis for improved anthrax vaccines. Adv. Drug Deliv. Rev. 2005;57:1266–1292. doi: 10.1016/j.addr.2005.01.028. [DOI] [PubMed] [Google Scholar]
- Brodsky FM, Guagliardi LE. The cell biology of antigen processing and presentation. Annu. Rev. Immunol. 1991;9:707–744. doi: 10.1146/annurev.iy.09.040191.003423. [DOI] [PubMed] [Google Scholar]
- Brossier F, Mock M. Toxins of Bacillus anthracis. Toxicon. 2001;39:1747–1755. doi: 10.1016/s0041-0101(01)00161-1. [DOI] [PubMed] [Google Scholar]
- Brossier F, Weber-Levy M, Mock M, Sirard JC. Role of toxin functional domains in anthrax pathogenesis. Infect. Immun. 2000;68:1781–1786. doi: 10.1128/iai.68.4.1781-1786.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham K, Lacy DB, Mogridge J, Collier RJ. Mapping the lethal factor and edema factor binding sites on oligomeric anthrax protective antigen. Proc. Natl. Acad. Sci. U S A. 2002;99:7049–7053. doi: 10.1073/pnas.062160399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedlander AM, Pittman PR, Parker GW. Anthrax vaccine: evidence for safety and efficacy against inhalational anthrax. Jama. 1999;282:2104–2106. doi: 10.1001/jama.282.22.2104. [DOI] [PubMed] [Google Scholar]
- Gorse GJ, Keitel W, Keyserling H, Taylor DN, Lock M, Alves K, Kenner J, Deans L, Gurwith M. Immunogenicity and tolerance of ascending doses of a recombinant protective antigen, rPA102. anthrax vaccine: a randomized, double-blinded, controlled, multicenter trial. Vaccine. 2006;24:5950–5959. doi: 10.1016/j.vaccine.2006.05.044. [DOI] [PubMed] [Google Scholar]
- Hammarstrom L, Carbonara AO, DeMarchi M, Lefranc G, Moller G, Smith CI, Zegers BJ. Subclass restriction pattern of antigen-specific antibodies in donors with defective expression of IgG or IgA subclass heavy chain constant region genes. Clin.Immunol. Immunopathol. 1987;45:461–470. doi: 10.1016/0090-1229(87)90097-3. [DOI] [PubMed] [Google Scholar]
- Jack HM, McDowell M, Steinberg CM, Wabl M. Looping out and deletion mechanism for the immunoglobulin heavy-chain class switch. Proc. Natl. Acad. Sci. U S A. 1988;85:1581–1585. doi: 10.1073/pnas.85.5.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabat EA, Wu TT, Perry HM, Gottesman KS, Foeller C. Sequences of Proteins of Immunological Interest. U.S. Department of Health and Human Services; Bethesda: 1991. [Google Scholar]
- Lefranc MP, Giudicelli V, Ginestoux C, Bodmer J, Muller W, Bontrop R, Lemaitre M, Malik A, Barbie V, Chaume D. IMGT, the international ImMunoGeneTics database. Nucleic Acids Res. 1999;27:209–212. doi: 10.1093/nar/27.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppla S. Anthrax Toxins. In: Moss J, editor. Baterial toxins and virulence factors in disease. Vol. 8. Marcel Dekker, Inc.; 1995. pp. 543–572. [Google Scholar]
- Leppla SH, Robbins JB, Schneerson R, Shiloach J. Development of an improved vaccine for anthrax. J. Clin. Invest. 2002;110:141–144. doi: 10.1172/JCI16204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little SF, Lowe JR. Location of receptor-binding region of protective antigen from Bacillus anthracis. Biochem. Biophys. Res. Commun. 1991;180:531–537. doi: 10.1016/s0006-291x(05)81097-6. [DOI] [PubMed] [Google Scholar]
- Lucas AH, Moulton KD, Tang VR, Reason DC. Combinatorial library cloning of human antibodies to Streptococcus pneumoniae capsular polysaccharides: variable region primary structures and evidence for somatic mutation of Fab fragments specific for capsular serotypes 6B, 14, and 23F. Infect. Immun. 2001;69:853–864. doi: 10.1128/IAI.69.2.853-864.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda F, Ishii K, Bourvagnet P, Kuma K, Hayashida H, Miyata T, Honjo T. The complete nucleotide sequence of the human immunoglobulin heavy chain variable region locus. J. Exp. Med. 1998;188:2151–2162. doi: 10.1084/jem.188.11.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C, Stedra J, Kelsoe G, Cerny J. Facultative role of germinal centers and T cells in the somatic diversification of IgVH genes. J. Exp. Med. 1995;181:1319–1331. doi: 10.1084/jem.181.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills FC, Thyphronitis G, Finkelman FD, Max EE. Ig mu-epsilon isotype switch in IL-4-treated human B lymphoblastoid cells. Evidence for a sequential switch. J.Immunol. 1992;149:1075–1085. [PubMed] [Google Scholar]
- Novak JM, Stein MP, Little SF, Leppla SH, Friedlander AM. Functional characterization of protease-treated Bacillus anthracis protective antigen. J. Biol. Chem. 1992;267:17186–17193. [PubMed] [Google Scholar]
- Papadea C, Check IJ. Human immunoglobulin G and immunoglobulin G subclasses: biochemical, genetic, and clinical aspects. Crit. Rev. Clin. Lab. Sci. 1989;27:27–58. doi: 10.3109/10408368909106589. [DOI] [PubMed] [Google Scholar]
- Petosa C, Collier RJ, Klimpel KR, Leppla SH, Liddington RC. Crystal structure of the anthrax toxin protective antigen. Nature. 1997;385:833–838. doi: 10.1038/385833a0. [DOI] [PubMed] [Google Scholar]
- Reason DC, Wagner TC, Lucas AH. Human Fab fragments specific for the Haemophilus influenzae b polysaccharide isolated from a bacteriophage combinatorial library use variable region gene combinations and express an idiotype that mirrors in vivo expression. Infect. Immun. 1997;65:261–266. doi: 10.1128/iai.65.1.261-266.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reason DC, Zhou J. Correlation of antigenic epitope and antibody gene usage in the human immune response to Streptococcus pneumoniae type 23F capsular polysaccharide. Clin Immunol. 2004;111:132–136. doi: 10.1016/j.clim.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Reason DC, Zhou J. Codon insertion and deletion functions as a somatic diversification mechanism in human antibody repertoires. Biol. Direct. 2006;1:24. doi: 10.1186/1745-6150-1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G, Quispe J, Leppla SH, Mitra AK. Large-scale structural changes accompany binding of lethal factor to anthrax protective antigen: a cryo-electron microscopic study. Structure. 2004;12:2059–2066. doi: 10.1016/j.str.2004.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scobie HM, Rainey GJ, Bradley KA, Young JA. Human capillary morphogenesis protein 2 functions as an anthrax toxin receptor. Proc. Natl. Acad. Sci. U S A. 2003;100:5170–5174. doi: 10.1073/pnas.0431098100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sever JL, Brenner AI, Gale AD, Lyle JM, Moulton LH, Ward BJ, West DJ. Safety of anthrax vaccine: an expanded review and evaluation of adverse events reported to the Vaccine Adverse Event Reporting System, VAERS. Pharmacoepidemiol. Drug Saf. 2004;13:825–840. doi: 10.1002/pds.936. [DOI] [PubMed] [Google Scholar]
- Sever JL, Brenner AI, Gale AD, Lyle JM, Moulton LH, West DJ. Safety of anthrax vaccine: a review by the Anthrax Vaccine Expert Committee, AVEC. of adverse events reported to the Vaccine Adverse Event Reporting System, VAERS. Pharmacoepidemiol. Drug Saf. 2002;11:189–202. doi: 10.1002/pds.712. [DOI] [PubMed] [Google Scholar]
- Singh Y, Klimpel KR, Quinn CP, Chaudhary VK, Leppla SH. The carboxyl-terminal end of protective antigen is required for receptor binding and anthrax toxin activity. J. Biol. Chem. 1991;266:15493–15497. [PubMed] [Google Scholar]
- Wagner SD, Neuberger MS. Somatic hypermutation of immunoglobulin genes. Annu. Rev. Immunol. 1996;14:441–457. doi: 10.1146/annurev.immunol.14.1.441. [DOI] [PubMed] [Google Scholar]
- Zhou J, Lottenbach KR, Barenkamp SJ, Lucas AH, Reason DC. Recurrent variable region gene usage and somatic mutation in the human antibody response to the capsular polysaccharide of Streptococcus pneumoniae type 23F. Infect. Immun. 2002;70:4083–4091. doi: 10.1128/IAI.70.8.4083-4091.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Lottenbach KR, Barenkamp SJ, Reason DC. Somatic hypermutation and diverse immunoglobulin gene usage in the human antibody response to the capsular polysaccharide of Streptococcus pneumoniae Type 6B. Infect. Immun. 2004;72:3505–3514. doi: 10.1128/IAI.72.6.3505-3514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]


