Abstract
Histone nuclear factor P (HiNF-P) activates histone H4 gene transcription at the G1/S phase transition upon association with its cyclin E/CDK2 responsive co-factor p220NPAT. Here we characterize the gene regulatory pathways that control the proliferation-related expression of HiNF-P. The HiNF-P locus contains a single TATA-less 0.6 kbp promoter with multiple phylogenetically conserved transcription factor recognition motifs. Transient reporter gene assays with HiNF-P promoter deletions show that there are at least three distinct activating regions (-387/-201, -201/-100 and -100/-1) that support maximal transcription. HiNF-P gene transcription is activated by SP1 through the -100/-1 domain and repressed by E2F1 through the -201/-100 domain. The multifunctional co-regulators CBP and p300 also stimulate HiNF-P gene transcription through the -201/-1 core promoter. Importantly, the HiNF-P promoter is activated by both HiNF-P and p220NPAT. This auto-regulatory activation is further enhanced by cyclin E and CDK2, while blocked by CDK inhibition (i.e., p57KIP2 p27KIP1, p21CIP). Thus, the HiNF-P gene is a key non-histone target of p220NPAT and HiNF-P. The dependence of HiNF-P gene transcription on cyclin E/CDK2/p220NPAT signaling defines a novel feed-forward loop that may sustain HiNF-P expression in proliferating cells to support the cell cycle regulated synthesis of histone H4 proteins.
Keywords: cell cycle, E2F1, SP1, p300, CBP
1. INTRODUCTION
Histone nuclear factor P (HiNF-P) is the key transcription factor regulating cell cycle dependent histone H4 gene expression at the G1/S phase transition (Holmes et al., 2005; Miele et al., 2005; Mitra et al., 2003; van Wijnen et al., 1992). The HiNF-P dependent induction of histone H4 gene transcription is functionally and temporally coupled to the assembly of newly replicated DNA into nucleosomes, whereby DNA associates with core histone octamers that each contains two copies of newly synthesized histone H4 proteins. Our group has shown that the transcriptional activity of HiNF-P is modulated through association with a cyclin E/CDK2 regulated co-activator (Miele et al., 2005; Mitra et al., 2003) that was previously characterized as p220NPAT (Ma et al., 2000; Wei et al., 2003; Zhao et al., 1998; Zhao et al., 2000). Consistent with their critical roles in histone H4 gene expression, both HiNF-P and p220NPAT are rate-limiting for cell cycle progression near the G1/S phase transition (Gao et al., 2003; Mitra et al., 2003; Ye et al., 2003).
HiNF-P is a Zn-finger transcription factor with an N-terminal DNA binding domain that contains nine canonical C2H2-motifs (Mitra et al., 2003). HiNF-P recognizes an unusually large consensus sequence within histone H4 genes (Aziz et al., 1998; Holmes et al., 2005; Stein et al., 1991; van Wijnen et al., 1991; van Wijnen et al., 1992) that links H4 gene transcription to the Cyclin E/CDK2/p220NPAT/HiNF-P pathway. HiNF-P is related to H4TF-2, a DNA binding protein that was not characterized beyond its molecular mass (Dailey et al., 1988; Mitra et al., 2003; van Wijnen et al., 1992), and was serendipitously re-discovered as methyl CpG Binding Protein 2 (MBD2) Zn-finger Interacting Factor (MZIF) in yeast two hybrid assays (Sekimata et al., 2001; Sekimata and Homma 2004). The C-terminal region of HiNF-P contains conserved motifs that may support interactions with different partner proteins, including p220NPAT and RNA processing components (Miele et al., 2005). The interaction of HiNF-P with p220NPAT converts HiNF-P into a potent activator (Miele et al., 2005) and increases the stability of p220NPAT (Medina et al., 2006).
HiNF-P DNA binding activity has been detected in a broad range of cells and tissues from different mammalian species (Aziz et al., 1998; Langdahl et al., 1997; Shakoori et al., 1995; van den Ent et al., 1993; van der Meijden et al., 1998; van Wijnen et al., 1991; van Wijnen et al., 1992), and HiNF-P binding sites in the promoters of H4 genes are only occupied in cells progressing through the cell cycle, as established by genomic DNaseI footprinting (Hovhannisyan et al., 2003; Pauli et al., 1987) and chromatin immunoprecipitations (Miele et al., 2005; Mitra et al., 2003). HiNF-P (MIZF) mRNA is ubiquitously expressed in a number of tissues and cells (Sekimata et al., 2001) and robustly expressed in human embryonic stem cells (Becker et al., 2006; Becker et al., 2007). While the downstream actions of HiNF-P as the master regulator of the histone H4 gene family have been investigated, the control of HiNF-P levels in proliferating cells remains to be elucidated.
In this study, we have characterized the mouse HiNF-P promoter to define pathways that transcriptionally regulate HiNF-P gene expression. We exogenously expressed candidate transcription factors that were identified by recognition motif analyses and assessed their functional effects in reporter gene assays using a set of HiNF-P promoter deletions. Our results show that the HiNF-P promoter is modulated by multiple factors (e.g., SP1, E2F1, and HiNF-P) and responds to three different co-activators including p220NPAT, p300 and CBP. Our findings define the HiNF-P gene as a non-histone target of the cyclin E/CDK2/p220NPAT signaling pathway. This pathway thus controls the overall levels and activity of HiNF-P to modulate expression of histone H4 genes in conjunction with DNA synthesis during S phase.
2. MATERIALS AND METHODS
2.1 Preparation of reporter gene constructs and expression vectors
To identify elements that regulate transcription of the mouse HiNF-P gene and to characterize its structure, we generated a panel of HiNF-P promoter segments fused to the luciferase reporter gene. Two mouse HiNF-P gene promoter fragments were generated by direct PCR-amplification from AB2.2 mouse genomic DNA. We obtained a ~3.6 kbp MluI-BglII fragment spanning the promoter and the first intron, and a 597 bp MluI-BglII fragment spanning the promoter only (Fig. 1) that were each inserted into pGL2-basic (Promega, Madison, WI). The amplification primers used for the generation of the two fragments were: ~3.6 kbp reverse BglII primer, 5′-gta gat ctG GCC TTT ACC TTA GAG GAA GAC; 597 bp reverse BglII primer, 5′-taa gat ctG CCA GCG CCG CTG GTT TGC CCA G and the shared forward MluI primer 5′-gta cgc gtA GAG CAT TTG TCT AGC AAC CAG.
Figure 1. Regulatory organization of the HiNF-P locus.
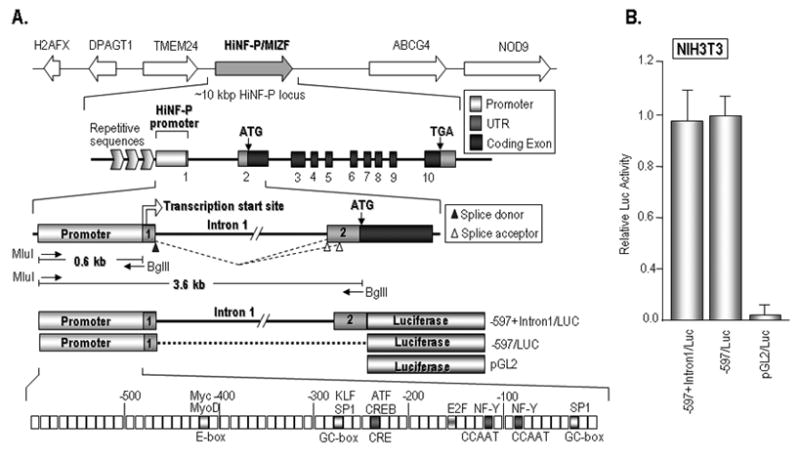
(A) Top: Chromosomal organization of the HiNF-P gene located on human chromosome 11 (11q23.3) and mouse chromosome 9. The mouse HiNF-P/MIZF is located ~5 kbp from the 3′ end of the TMEM24 gene. Middle: Enlargement of the ~10 kbp spanning the HiNF-P locus including coding exons, as well as 5′ and 3′ end regions. The first 0.6 kbp of the 5′ region of the HiNF-P gene contains unique sequences that are conserved in vertebrate species and represent the HiNF-P promoter. The 5′ border of the HiNF-P promoter is defined by repetitive sequences (horizontal chevrons) in the intergenic region shared with the TMEM24 locus. Bottom: DNA segments containing the mouse HiNF-P promoter (597 bp) or the promoter plus intron 1 (~3.6 kbp) were amplified by PCR using primers (small arrows) with the indicated restriction sites. These DNA fragments were fused to the luciferase gene to construct reporter gene vectors used in transfection assays. Bioinformatics analyses revealed consensus motifs for multiple transcription factors. (B) Reporter gene assays with the -597+Intron1/LUC and -597/LUC promoter/reporter gene constructs in comparison to the promoterless parent vector (pGL2) were performed in NIH3T3 cells. The graph shows average luciferase values for a representative experiment (error bars show standard deviation, n=3). Similar results were obtained in three independent replicates in triplicate.
To delineate the elements responding to multiple transcription factors, we prepared four additional mouse HiNF-P promoter deletion constructs by PCR, designated -382 HiNF-P/Luc, -201 HiNF-P/Luc, -100 HiNF-P/Luc and -80 HiNF-P/Luc using -597 HiNF-P/Luc as a template. The -597 reverse/BglII primer (see above) was used with the following forward primers to prepare the first three constructs: -382 forward MluI primer, 5′-aac acg cgt TGG GGA AGA TGA CAG ACA AGG-3′;-201 forward MluI primer, 5′-gaa acg cgt TCC CCC TTG GGA CTC GGA G-3′;-100 forward MluI primer, 5′-aagacgcgt GAC TCG AGA GCG GCG ATT GG-3′. The -80 HiNF-P/Luc construct was generated by a synthetic MluI/BglII fragment of two complementary oligonucleotides: 5′-cgc gtCT GTC GCT GGT CGT CGC TCA AAC GCC TCG GGG TAG ATG GTG TGA GGT GGG TGG GGC GCT GGG CAA ACC AGC GGC GCT GGC a-3′; 5′-gat ctG CCA GCG CCG CTG GTT TGC CCA GCG CCC CAC CCA CCT CAC ACC ATC TAC CCC GAG GCG TTT GAG CGA CGA CCA GCG ACA Ga-3′; both oligonucleotides were directly inserted into the MluI/BglII site of pGL2-basic. All oligonucleotides were synthesized using a 1000M DNA synthesizer (Beckman). All constructs were subject to automated sequencing to verify the correct orientation of promoter segments relative to the reporter gene and absence of polymerase chain reaction or chemical synthesis-related mutations. We thank Drs. Guntram Suske (Philipps-University Marburg, Germany) and Wade Harper (Harvard Medical School, Boston, MA) for kindly providing CMV driven expression constructs for SP1 and p220NPAT.
2.2 Cell culture, transfection experiments, luciferase reporter assays and western blot analysis
Actively proliferating cultures of NIH3T3, HeLa and WI38 cells were maintained in Dulbecco’s modified Eagle’s medium (Life Technologies), MC3T3 cells were maintained in α MEM medium, and SAOS-2 cells were maintained in McCoy’s 5A medium supplemented with 10% fetal calf serum (or 15%FCS for SAOS-2 cells), 100 units/ml penicillin, 100 μg/ml streptomycin (Sigma) and 2 mM L-glutamine (Sigma) at 37°C in humidified air containing 5% CO2. Cells were seeded in 6-well culture plates (Greiner) at a density of 1.5–2.0 × 105 cells/well, and transient transfections were performed 24 h later using the Superfect transfection method (Qiagen) or Fugene 6 method (Roche). We co-transfected 0.1 μg/well of each mHiNF-P promoter/luciferase reporter gene construct with 0.15 μg/well of each expression gene vector. The total amount of DNA in each well was kept constant by supplementing the transfection mixture with the empty expression vector. Lysates for use in luciferase assays were prepared from cells harvested 24 hr after transfection. Each transfection experiment was performed in triplicate and each experiment was repeated at least three times.
Cells were washed twice with 1X PBS buffer at the time of harvest, and lysed with 1X lysis buffer (Promega). Luciferase assays were carried out according to the specifications of the manufacturer using a standard luciferase reporter assay system (Promega), and luminometric units were determined by a Monolight 2010 Luminometer (Analytical Luminescence Laboratory). The same protein samples as used in luciferase assays were subjected to western blotting following 4–15 % Linear Gradient SDS-PAGE (Bio-RAD). Upon transfer, membranes were probed with antibodies against the following proteins: HiNF-P (rabbit IgG); p220 (BD Bioscience); E2F1, SP1, p300 and CBP (all from Santa Cruz, CA).
3. RESULTS
3.1 Definition of the promoter that controls transcription of the mouse HiNF-P locus
To understand regulation of the mouse HiNF-P gene, we cloned the HiNF-P promoter based on available sequence data of the murine genome. The HiNF-P locus is located on human chromosome 11q23.3 and mouse chromosome 9 in a syntenic region in which the genes for H2AX, DPAGT1, TMEM24, HiNF-P and ABCG4 are organized in the same order within a 0.1 Mbp segment (Fig. 1A). The HiNF-P gene encompasses 10 exons, and transcription initiates with Exon 1, a mini-exon that encodes only ~30 bp of 5′UTR sequences of the mature HiNF-P mRNA. Exon 1 splices into Exon 2 which contains the translational start codon of the HiNF-P protein. The TMEM24 gene is located about 5 kbp upstream from the HiNF-P gene and is transcribed in the same orientation. The 3′ end of the TMEM24 gene and 5′ end of the HiNF-P gene are separated by a series of repetitive sequences that terminate approximately 0.6 kB from the transcriptional start site of the HiNF-P gene as defined by the endpoints of full length mouse and human cDNAs. The proximity of the TMEM24 gene, which encodes a transmembrane protein with a biologically distinct function, suggests that the key elements controlling HiNF-P gene transcription may be concentrated close to the transcriptional start site of the HiNF-P mRNA. The repetitive sequences beyond 0.6 kbp of the HiNF-P 5′ region represent a natural demarcation of the distal boundary of the HiNF-P promoter. The HiNF-P promoter contains several recognition motifs for canonical transcription factors (e.g., inverted CCAAT/NF-Y boxes, GC-boxes, ATF sites) that are conserved among multiple mammalian species (e.g., human, chimpanzee, mouse, rat, dog, and cow; data not shown), as well as a number of CpG doublets, but does not appear to have a TATA-box.
3.2 HiNF-P transcription is controlled by multiple gene regulatory elements
We initially assessed the activity of the 0.6 kbp HiNF-P promoter in the presence or absence of the ~3 kbp region that encompasses Intron 1, to examine the formal possibility that this intron might harbor regulatory information (e.g., enhancer elements) that could influence HiNF-P transcription. Reporter gene constructs containing the mouse HiNF-P promoter without (-597 HiNF-P/LUC) or with Intron 1 (-597+Intron 1 HiNF-P/LUC) were transiently transfected into mouse NIH3T3 cells (Fig. 1B). The data clearly show that the -579 bp HiNF-P promoter supports a robust level of basal transcription that is significantly elevated above background levels observed for a promoter-less LUC construct (pGL2/LUC)(Fig. 1B). Furthermore, similar levels of basal activation are observed in the presence or absence of Intron 1 sequences. Hence, the 0.6 kbp promoter suffices for high level transcription of HiNF-P mRNA.
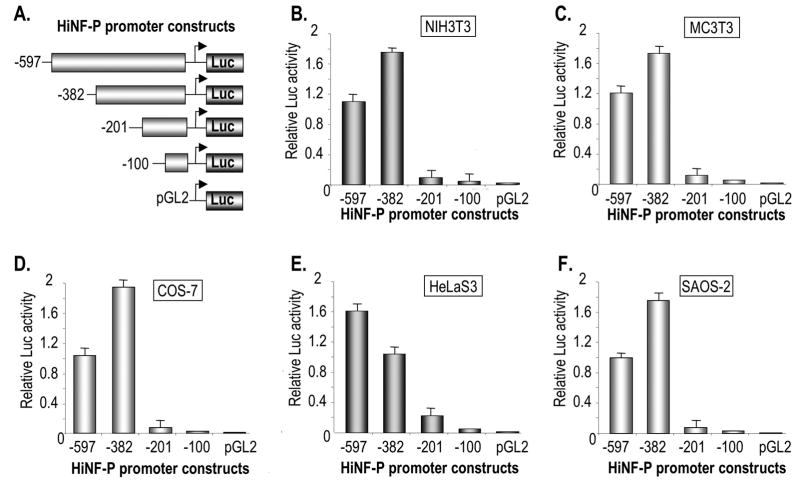
To delineate the elements that support transcription of the HiNF-P gene, we generated a series of specific deletions within the promoter of the HiNF-P gene (i.e., -597/LUC, -382/LUC, -201/LUC, -100/LUC) that are fused to the LUC reporter gene in the pGL2 construct. The resulting HiNF-P promoter deletion constructs were tested in a panel of cell types (SAOS-2, HeLa S3, COS-7, NIH3T3, and MC3T3 cells). The results show that the most pronounced change in basal promoter activity occurs upon deletion of the -382/-201 region with additional reductions in transcription observed upon deletion of HiNF-P promoter segments -201/-100 and -100/-1 (Fig. 2). Deletion of each of these segments reduces promoter activity similarly in all cell types examined. In contrast, deletion of the region between -597 to -382 appears to enhance promoter activity in most cell types, but reduces transcription by ~30% in HeLa S3 cells. Thus, the first 0.6 kbp of the HiNF-P promoter comprises several regulatory domains.
Figure 2. Delineation of elements controlling HiNF-P transcription.
(A) Diagram of HiNF-P promoter/luciferase reporter gene constructs used in this study with the indicated lengths of 5′ region. The reporter gene constructs (100 ng) were transiently transfected in triplicate into NIH3T3 (B), MC3T3 (C), COS-7 (D), HeLaS3 (E), or SAOS-2 (F) cells seeded in 6-well plates. The graphs show the average luciferase activity after 36 hr of a representative triplicate experiment (error bars show standard deviation) that was independently repeated three times.
3.3 Activation and co-activation of the HiNF-P promoter by distinct gene regulatory factors
Bioinformatics analysis and visual inspection of the HiNF-P promoter sequence revealed a number of putative recognition motifs for transcription factors and elements that respond to co-activating proteins (Fig. 1A and data not shown). To address the functional significance of these putative regulatory elements, we transiently expressed a panel of distinct sequence-specific transcription factors (e.g., SP1, E2F1 and HiNF-P) or co-activators (i.e., p300, CBP and p220NPAT) in parallel transfections with the -597/LUC reporter (Fig. 3). These experiments were also performed with the -597+Intron 1/LUC reporter and yielded very similar results (data not shown). For each transfected factor, we assessed quantitative effects on HiNF-P promoter activity, as well as the level of exogenous expression (Fig. 3 and data not shown). Western blot results show that each of the six proteins (i.e., HiNF-P, SP1, E2F1, p300, CBP and p220NPAT) is expressed at levels several-fold higher than the corresponding endogenous level (Fig. 3B). The data show that SP1 is a strong activator of the HiNF-P gene, that HiNF-P can modestly stimulate transcription of its own gene, and that E2F1 strongly represses transcription in SAOS-2 cells (Fig. 3A). Furthermore, each of the three co-activators (i.e., p300, CBP and p220NPAT) stimulated HiNF-P promoter activity. Hence, the activity of the HiNF-P promoter is controlled by multiple factors.
Figure 3. Trans-activation of the HiNF-P Promoter by multiple gene regulatory factors.
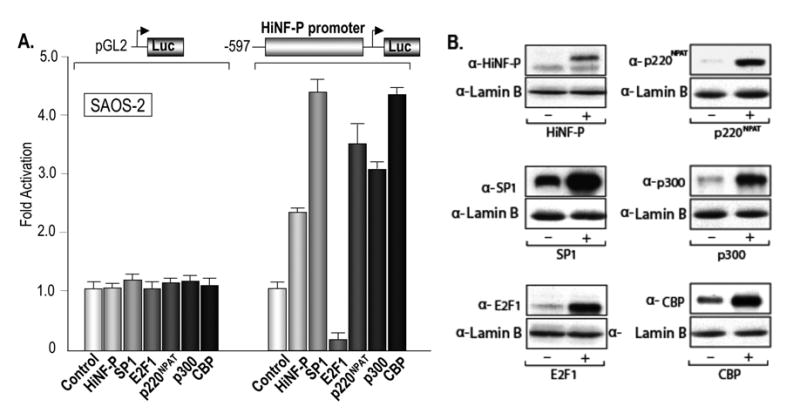
(A) The bar graph shows luciferase results obtained upon co-transfection of the full length HiNF-P promoter/Luciferase construct and expression vectors for the gene regulatory factors HiNF-P, SP1, E2F, p220NPAT, p300 and CBP as indicated. The reporter gene constructs (100 ng) and expression vector (200 ng) were transiently transfected in triplicate into SAOS-2 cells. (B) Western blot analysis of luciferase cell lysates for each of the factors expressed in Panel A (+; right lanes) relative to lysates from control cells transfected with empty expression vectors (−; left lanes). Each lysate was analyzed with the indicated antibodies to detect endogenous versus exogenous expression levels of HiNF-P, SP1, E2F, p220NPAT, p300 or CBP. The nuclear protein Lamin B was used as an internal control for protein quantitation.
3.4 Co-stimulation by p220NPAT signaling and suppression by E2F1 signaling
We determined the effects of co-transfecting the set of factors controlling the HiNF-P promoter in the presence of p220NPAT or E2F1 in SAOS-2 cells. The data reveal that p220NPAT is capable of co-stimulating HiNF-P promoter activity together with its partner protein HiNF-P, as well as with SP1, p300 and CBP (Fig. 4A). However, p220NPAT can not stimulate transcription effectively (i.e., above basal levels observed in the controls) in the presence of E2F-1 (Fig. 4A). In comparison, E2F-1 is an effective antagonist of the set of five factors that stimulate HiNF-P promoter activity in SAOS-2 cells. For each factor tested, E2F1 reduces promoter activity below the control value of cells not expressing exogenous factors (Fig. 4B). Hence, while the p220NPAT pathway generates a positive transcriptional response, E2F1 is a potent suppressor of HiNF-P gene transcription.
Figure 4. Synergism involving p220NPAT and antagonism by E2F1 at the HiNF-P promoter.
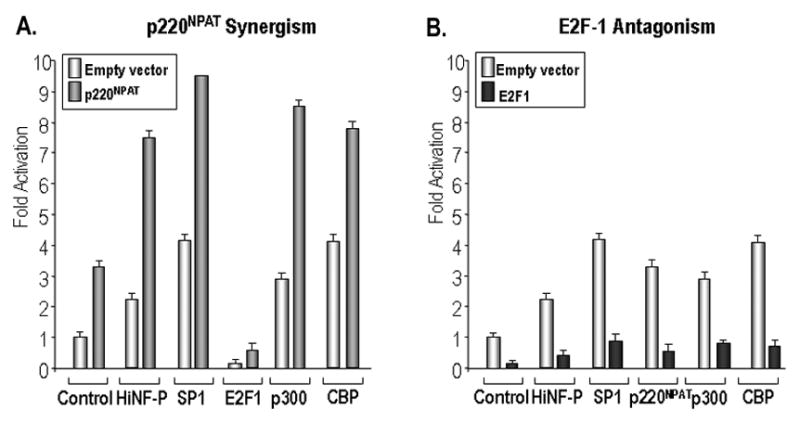
Reporter gene assays were performed in SAOS-2 cells with the full length HiNF-P promoter (100 ng) to examine synergism with p220NPAT (A) and antagonism by E2F1 (B). Each protein was co-expressed in combination with a second expression vector as indicated below the bar graphs. The set of two control transfections contains empty vector (100 ng; left) or expression vector for p220NPAT or E2F1 (100 ng; right). The total DNA amount of expression vector was maintained at 200 ng in all transfections. Luciferase values were normalized to control cells transfected with empty vectors.
3.5 Delineation of elements responding to activators and co-activators of the HiNF-P promoter
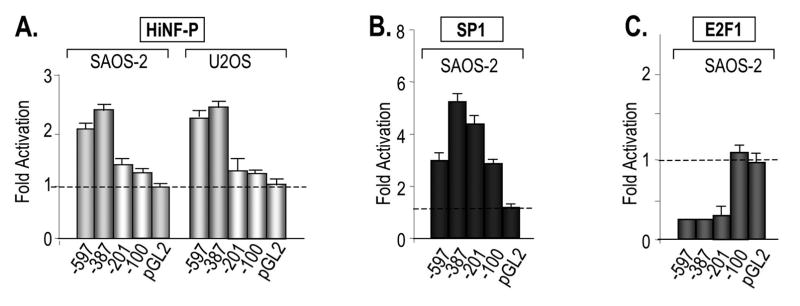
To delineate the regions in the HiNF-P promoter responding to each factor, we performed co-expression studies with our panel of HiNF-P promoter deletions using several different cell types (e.g., SAOS-2, U2OS, HeLa S3, WI38, and/or NIH3T3) (Fig. 5 and data not shown). Co-expression of HiNF-P increases the activity of both the -597 and -387 promoters, but not the -201 and -100 promoters (Fig. 5A). These results indicate that HiNF-P controls the promoter of its own gene and that this auto-activation requires the -387/-201 segment. SP1 can activate all HiNF-P promoter segments tested, including the -100 promoter. SP1 promoter activation is only marginally reduced when the -387/-201 segment is deleted, but completely abolished upon removal of the -100/-1 segment. These data indicate that the consensus SP1 element (i.e., GC-box) in the -100/-1 promoter region is sufficient for SP1 dependent activation (see Fig. 1A). SP1 activation of the HiNF-P promoter was observed in all cell types examined (i.e., SAOS-2, HeLaS3 and WI-38) albeit that the extent of stimulation differs among cell types from ~2 to 6 fold (Fig. 5B and data not shown). E2F1 represses HiNF-P promoter deletions -387 and -201, but not -100, indicating that E2F1 repression in SAOS-2 cells occurs through the -201/-100 region (Fig. 5C). Consistent with our results, the -201/-100 region contains a canonical E2F-1 binding site (Fig. 1). Taken together, the data establish that HiNF-P (-387/-201), E2F1 (-201/-100) and SP1 (-201/-100 and -100/-1) modulate HiNF-P promoter activity through at least three distinct 5′ regulatory elements.
Figure 5. Delineation of HiNF-P promoter elements responding to HiNF-P, SP1 and E2F.
The graphs show reporter gene assays that were performed with luciferase constructs containing different deletions of the HiNF-P promoter and expression vectors for HiNF-P (A), SP1 (B) and E2F1 (C). The indicated gene regulatory factors that were co-transfected into SAOS-2 and U2OS cells (see Figures 2 and 3 for other details). Luciferase values were normalized to the respective controls (i.e., transfections with corresponding empty vectors) which were set as one to emphasize the fold activation by each factor. Each HiNF-P promoter deletion construct has a distinct basal transcription level as is shown in Figure 2.
To determine the regions of the HiNF-P promoter that are controlled by the transcriptional co-activators p220NPAT, p300 and CBP, we carried out co-transfections with the same panel of HiNF-P promoter deletions and expression vectors for each of the three co-factors. There are two elements (i.e., -387/-201 and -100/-1) that modulate the response to elevations in the levels of p220NPAT (Fig. 6A). Stimulation of the HiNF-P promoter by the multifunctional co-factor p300 (Fig. 5) in SAOS-2 cells occurs through the -201/-100 region, while the response element for the histone acetyl transferase CBP maps to the -100/-1 region (Fig. 6B). Thus, the three co-factors we tested regulate transcription through three different HiNF-P promoter segments (Fig. 6C).
Figure 6. Delineation of HiNF-P promoter elements responding to p220NPAT, p300 and CBP.
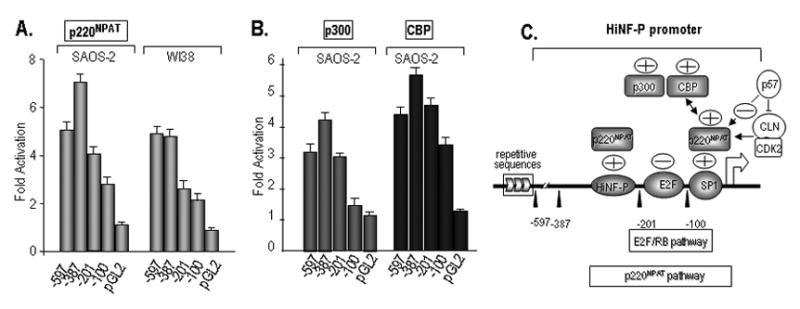
Transcription assays were performed with SAOS-2 and WI-38 cells as described in Figure 5 using a panel of Luciferase constructs (100 ng) fused to the indicated HiNF-P promoter deletions and expression vectors (200 ng each) for p220NPAT (A), p300 and CBP (B). Luciferase values were normalized to the respective controls which were set as one. (C) Pathways converging on the HiNF-P promoter identified in this study are indicated. The proximal promoter responds to the levels of SP1 and E2F that each have known consensus motifs. The HiNF-P promoter is also stimulated by the co-activators p300 and CBP that are known to control histone acetylation. Both proteins may interact with the HiNF-P promoter through p220NPAT. HiNF-P also enhances the activity of its own promoter but motifs for HiNF-P are not readily apparent in the promoter.
3.6 The HiNF-P promoter responds to the Cyclin E/CDK2/p220NPAT signaling pathway
The HiNF-P co-factor p220NPAT was initially characterized as a cyclin E/CDK2 substrate (Zhao et al., 1998). Because we observe that p220NPAT is capable of stimulating the -100/-1 HiNF-P promoter (see Fig. 6), we examined whether forced expression of CDK2 and cyclin E can enhance this stimulation. Co-expression of only CDK2 increases activity of the -100/-1 promoter, but not the promoter-less construct (pGL2-LUC), while forced expression of only Cyclin E does not have appreciable effects (Fig. 7A). Combined expression of p220NPAT, cyclin E and CDK2 results in strong synergistic activation of the HiNF-P promoter (Fig. 7A).
Figure 7. Synergistic activation of the HiNF-P promoter by the Cyclin E/CDK2/p220NPAT pathway and inhibition by p57KIP2.
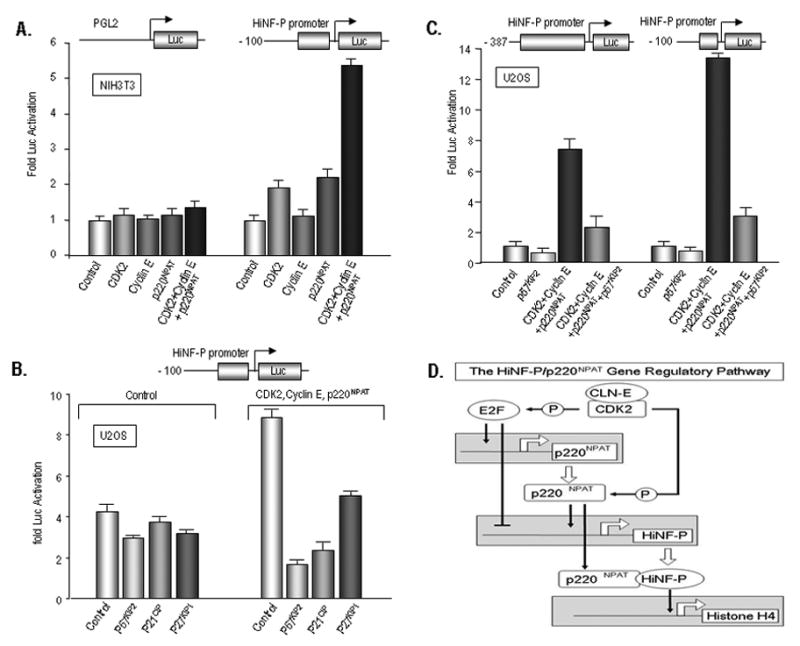
(A) Reporter gene assays were performed with the -100 HiNF-P promoter deletion construct (100 ng) in the presence of expression vectors for CDK2 (50 ng), cyclin E (50 ng), p220NPAT (100 ng) or the combination of all proteins in NIH3T3 cells. Control transfections were performed with empty vector (200 ng) and empty vector was included in the remaining reactions to maintain the total DNA amount of expression vector at 200 ng. Luciferase values were normalized to the respective controls (i.e., transfections with empty vectors) which were set as one to emphasize the fold activation by each factor. (B) Reporter gene assays were performed with the -100 HiNF-P promoter deletion construct (100 ng) or the promoter-less Luc construct in the presence of expression vectors for p21CIP, p27KIP1, or p57KIP2 (50 ng) in U2OS cells. (C) Reporter gene assays were performed with the -397 promoter or the -100 HiNF-P promoter (100 ng) in the presence of expression vectors for CDK2 (50 ng), cyclin E (50 ng), p220NPAT (100 ng) or the combination of all proteins in the absence or presence of p57KIP2 (50 ng) in U2OS cells. (D) Novel cell cycle regulatory feed-back loops may control the G1/S phase transition. HiNF-P and its co-activator p220NPAT stimulate the HiNF-P gene promoter, while both proteins interact to induce the cell cycle dependent induction of histone H4 expression during S phase. Both the p220NPAT and HiNF-P gene promoters are modulated by E2F1, while both p220NPAT (as a direct substrate) and E2F1 (through inactivation of pocket proteins) are stimulated by cyclin E/CDK2 signaling. Another level of autoregulation is provided by trans-activation of the p220NPAT promoter by HiNF-P and stabilization of p220NPAT by HiNF-P through protein/protein interactions (Medina et al., 2006).
The CDK inhibitory (CKI) proteins p21CIP, p27KIP1 and p57KIP2 can all block CDK2 activity. Therefore, we examined whether these three CKIs can alter HiNF-P promoter activity. Luciferase assays clearly show that p21CIP, p27KIP1 and p57KIP2 all have limited suppressive effects on the basal activity of the -100/-1 HiNF-P promoter. However, all three CKIs inhibit the stimulation of HiNF-P gene transcription by p220NPAT, CDK2 and cyclin E (Fig. 7B), with p57KIP2 representing the most potent inhibitor.
Because p220NPAT can control HiNF-P promoter activity through two elements in either the -100/-1 or -387/-201 promoter, we compared co-activation by CDK2, Cyclin E and p220NPAT together on both promoters. Our data show that both promoter segments are capable of transducing the Cyclin E/CDK2/p220NPAT signaling pathway and mediate strong transcriptional enhancement (Fig. 7C). Although the -100 promoter appears to be more effectively stimulated than the -387 promoter, the -100 promoter also has considerably lower basal activity (see Fig. 2). The -387 promoter is most highly transcribed when stimulated by the Cyclin E/CDK2/p220NPAT signaling pathway. The increase in activity of the -387 and -100 HiNF-P promoters is blocked in both cases by p57KIP2 (as a representative CKI). We conclude that regulation of HiNF-P by p220NPAT provides an autoregulatory feed forward loop that may support HiNF-P gene expression during the cell cycle (Fig. 7D).
4. DISCUSSION
The DNA binding activity of HiNF-P that supports its interaction with the genomic loci of histone H4 genes in vivo (Hovhannisyan et al., 2003; Pauli et al., 1987) is specific for a broad spectrum of proliferating cells (Aziz et al., 1998; Mitra et al., 2003; Shakoori et al., 1995; van den Ent et al., 1993; van Wijnen et al., 1991; van Wijnen et al., 1992). HiNF-P protein levels and in vivo occupancy of its site are constitutive during the cell cycle (Medina et al., 2006; Pauli et al., 1987), but both DNA binding activity and protein levels of HiNF-P are down regulated below the level of detection upon differentiation of HL60 cells (Hovhannisyan et al., 2003). The mRNA for HiNF-P is expressed in human embryonic stem cells (Becker et al., 2006; Becker et al., 2007), as well as in many distinct mammalian somatic cells and tissues (Mitra et al., 2003; Sekimata et al., 2001). Furthermore, our examination of expression profiling data deposited in the Gene Expression Omnibus (GEO) and the Expressed Sequence Tag (EST) databases for HiNF-P/MIZF indicates that its mRNA is expressed at the earliest stages of vertebrate embryogenesis, including mouse oocytes and 1-cell stage embryos (GEO accession # GDS2387; data deposited by Keith Latham, Temple University), as well as mouse day 3 blastocysts (our unpublished observations). In this study, we examined the gene regulatory pathways that converge on the HiNF-P locus to control its proliferation-specific expression.
Our data show that transcription of the HiNF-P gene is controlled by a TATA-less core promoter spanning ~0.4 kbp and containing at least three distinct cis-acting regions (-387/-201, -201/-100 and -100/-1) that are responsive to several different trans-acting regulatory pathways. The apparent compactness of the HiNF-P gene promoter is comparable to that of histone genes in which the essential regulatory information is confined to small proximal 5′ regulatory sequences. The condensation of regulatory information near the transcriptional start site facilitates evolutionary conservation during recombination events, perhaps reflected by location of the HiNF-P gene within a syntenic chromosomal region in the mouse, rat and human genomes. Because activity of the HiNF-P/p220NPAT co-activation complex is essential during the cell cycle and development (Di Fruscio et al., 1997; Miele et al., 2005; Mitra et al., 2003; Ye et al., 2003), the HiNF-P gene must retain its critical regulatory elements to ensure appropriate expression.
We establish that HiNF-P gene transcription is activated by the ubiquitous G/C box binding protein SP1 through at least one element in the -100/-1 region. A second element located in the 387/-201 region did not appear to be rate-limiting for transcription; this second element may represent a low affinity SP1 binding site and/or the functional element in the -100/-1 region may compensate for the deletion of the SP1 site in the -387/-200 region. SP1 regulates house-keeping genes under control of TATA-less promoters (Suske 1999), as well as cell cycle related genes (e.g., histone H4 and CDK2) (Birnbaum et al., 1995; Xie et al., 2003). Control of the HiNF-P gene promoter by SP1 may account at least in part for the proliferation-specific expression of HiNF-P that we have observed in many cell types (Aziz et al., 1998; Gruol and Altschmied 1993; Hovhannisyan et al., 2003; Mitra et al., 2003; Shakoori et al., 1995; van der Meijden et al., 1998; van Wijnen et al., 1991). We also find that the E2F1 represses the HiNF-P gene in SAOS-2 cells. Because E2F1 normally controls S phase related genes positively, the finding that E2F1 negatively regulates HiNF-P gene transcription is unexpected. Our co-expression experiments reveal that E2F1 and SP1 negate each others opposing effects. Thus, the balance of these two factors may establish the basal mRNA levels of HiNF-P in rapidly dividing cells.
One principal finding of this study is that the HiNF-P promoter is transcriptionally stimulated by HiNF-P and p220NPAT, as well as co-stimulated by cyclin E/CDK2, but inhibited by a CDK inhibitor (i.e., p57KIP2, p27KIP1 p21CIP. Furthermore, HiNF-P promoter activity is also stimulated by the multifunctional co-regulators CBP and p300. Recent studies suggest that these co-regulators may interact with p220NPAT to control transcription in S phase (Wang et al., 2004). Thus, the transcriptional enhancement of the HiNF-P gene by CBP and p300 may be directly related to our observation that the HiNF-P promoter responds to p220NPAT. We have recently shown that HiNF-P stabilizes p220NPAT through protein/protein interactions (Medina et al., 2006), indicating that the two principal components of the HiNF-P/p220NPAT co-activation complex are self-regulated at transcriptional and/or post-transcriptional levels. Taken together, our findings suggest that activation of the cyclin E/CDK2/p220NPAT/HiNF-P signaling pathway generates a self-sustaining feed-forward loop to induce cell cycle controlled histone H4 gene expression in proliferating cells.
Acknowledgments
The work was supported by National Institutes of Health grant GM032010 and human stem cell supplement. We thank Guntram Suske (Philipps-University Marburg, Germany), Wade Harper (Harvard Medical School, Boston, MA) and Scott Hiebert (Vanderbilt University) for kindly providing reagents. We also thank the members of our research group, and especially Ricardo Medina, Prachi Gupta and Margaretha van der Deen, for stimulating discussions.
Abbreviations
- HiNF-P
Histone Nuclear Factor P
- CDK
cyclin dependent kinase
- CIP
CDK Inhibitory Protein
- CKI
CDK Inhibitory Protein
- KIP
cyclin dependent Kinase Inhibitory Protein
- NPAT
nuclear protein at ataxia telangiectasia locus
- E2F1
adenovirus early gene transcription factor 1
- SP1
stimulatory protein 1
- CREB
Cyclic AMP Responsive Element Binding protein
- CBP
CREB Binding Protein
- H4TF-2
H4 Transcription Factor 2
- MBD2
Methyl CpG Binding Domain protein 2
- MIZF
MBD2 Interaction Zinc Finger protein
- Luc
luciferase
- MEM
Eagle’s minimal essential medium
- EST
expressed sequence tag
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aziz F, van Wijnen AJ, Vaughan PS, Wu S, Shakoori AR, Lian JB, Soprano KJ, Stein JL, Stein GS. The integrated activities of IRF-2 (HiNF-M) CDP/cut (HiNF-D) and H4TF-2 (HiNF-P) regulate transcription of a cell cycle controlled human histone H4 gene: mechanistic differences between distinct H4 genes. Mol Biol Rep. 1998;25:1–12. doi: 10.1023/a:1006888731301. [DOI] [PubMed] [Google Scholar]
- Becker KA, Ghule PN, Therrien JA, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Self-renewal of human embryonic stem cells is supported by a shortened G1 cell cycle phase. J Cell Physiol. 2006;209:883–893. doi: 10.1002/jcp.20776. [DOI] [PubMed] [Google Scholar]
- Becker KA, Stein JL, Lian JB, van Wijnen AJ, Stein GS. Establishment of histone gene regulation and cell cycle checkpoint control in human embryonic stem cells. J Cell Physiol. 2007;210:517–526. doi: 10.1002/jcp.20903. [DOI] [PubMed] [Google Scholar]
- Birnbaum MJ, van Wijnen AJ, Odgren PR, Last TJ, Suske G, Stein GS, Stein JL. Sp1 trans-activation of cell cycle regulated promoters is selectively repressed by Sp3. Biochemistry. 1995;34:16503–16508. doi: 10.1021/bi00050a034. [DOI] [PubMed] [Google Scholar]
- Dailey L, Roberts SB, Heintz N. Purification of the human histone H4 gene-specific transcription factors H4TF-1 and H4TF-2. Genes Dev. 1988;2:1700–1712. doi: 10.1101/gad.2.12b.1700. [DOI] [PubMed] [Google Scholar]
- Di Fruscio M, Weiher H, Vanderhyden BC, Imai T, Shiomi T, Hori TA, Jaenisch R, Gray DA. Proviral inactivation of the Npat gene of Mpv 20 mice results in early embryonic arrest. Mol Cell Biol. 1997;17:4080–4086. doi: 10.1128/mcb.17.7.4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G, Bracken AP, Burkard K, Pasini D, Classon M, Attwooll C, Sagara M, Imai T, Helin K, Zhao J. NPAT expression is regulated by E2F and is essential for cell cycle progression. Mol Cell Biol. 2003;23:2821–2833. doi: 10.1128/MCB.23.8.2821-2833.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruol DJ, Altschmied J. Synergistic induction of apoptosis with glucocorticoids and 3′, 5′-cyclic adenosine monophosphate reveals agonist activity by RU 486. Mol Endocrinol. 1993;7:104–113. doi: 10.1210/mend.7.1.8383286. [DOI] [PubMed] [Google Scholar]
- Holmes WF, Braastad CD, Mitra P, Hampe C, Doenecke D, Albig W, Stein JL, van Wijnen AJ, Stein GS. Coordinate control and selective expression of the full complement of replication-dependent histone H4 genes in normal and cancer cells. J Biol Chem. 2005;280:37400–37407. doi: 10.1074/jbc.M506995200. [DOI] [PubMed] [Google Scholar]
- Hovhannisyan H, Cho B, Mitra P, Montecino M, Stein GS, van Wijnen AJ, Stein JL. Maintenance of open chromatin and selective genomic occupancy at the cell-cycle-regulated histone H4 promoter during differentiation of HL-60 promyelocytic leukemia cells. Mol Cell Biol. 2003;23:1460–1469. doi: 10.1128/MCB.23.4.1460-1469.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdahl BL, Knudsen JY, Jensen HK, Gregersen N, Eriksen EF. A sequence variation: 713-8delC in the transforming growth factor-beta 1 gene has higher prevalence in osteoporotic women than in normal women and is associated with very low bone mass in osteoporotic women and increased bone turnover in both osteoporotic and normal women. Bone. 1997;20:289–294. doi: 10.1016/s8756-3282(96)00363-8. [DOI] [PubMed] [Google Scholar]
- Ma T, Van Tine BA, Wei Y, Garrett MD, Nelson D, Adams PD, Wang J, Qin J, Chow LT, Harper JW. Cell cycle-regulated phosphorylation of p220(NPAT) by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev. 2000;14:2298–2313. doi: 10.1101/gad.829500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina R, van Wijnen AJ, Stein GS, Stein JL. The histone gene transcription factor HiNF-P stabilizes its cell cycle regulatory co-activator p220NPAT. Biochemistry. 2006;45:15915–15920. doi: 10.1021/bi061425m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele A, Braastad CD, Holmes WF, Mitra P, Medina R, Xie R, Zaidi SK, Ye X, Wei Y, Harper JW, van Wijnen AJ, Stein JL, Stein GS. HiNF-P directly links the cyclin E/CDK1/p220NPAT pathway to histone H4 gene regulation at the G1/S phase cell cycle transition. Mol Cell Biol. 2005;25:6140–6153. doi: 10.1128/MCB.25.14.6140-6153.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra P, Xie RL, Medina R, Hovhannisyan H, Zaidi SK, Wei Y, Harper JW, Stein JL, van Wijnen AJ, Stein GS. Identification of HiNF-P, a key activator of cell cycle controlled histone H4 genes at the onset of S phase. Mol Cell Biol. 2003;23:8110–8123. doi: 10.1128/MCB.23.22.8110-8123.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli U, Chrysogelos S, Stein G, Stein J, Nick H. Protein-DNA interactions in vivo upstream of a cell cycle- regulated human H4 histone gene. Science. 1987;236:1308–1311. doi: 10.1126/science.3035717. [DOI] [PubMed] [Google Scholar]
- Sekimata M, Homma Y. Sequence-specific transcriptional repression by an MBD2-interacting zinc finger protein MIZF. Nucleic Acids Res. 2004;32:590–597. doi: 10.1093/nar/gkh249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekimata M, Takahashi A, Murakami-Sekimata A, Homma Y. Involvement of a novel zinc finger protein, MIZF, in transcriptional repression by interacting with a methyl-CpG-binding protein, MBD2. J Biol Chem. 2001;276:42632–42638. doi: 10.1074/jbc.M107048200. [DOI] [PubMed] [Google Scholar]
- Shakoori AR, van Wijnen AJ, Cooper C, Aziz F, Birnbaum M, Reddy GP, Grana X, De Luca A, Giordano A, Lian JB, Stein JL, Quesenberry P, Stein GS. Cytokine induction of proliferation and expression of CDC2 and cyclin A in FDC-P1 myeloid hematopoietic progenitor cells: regulation of ubiquitous and cell cycle-dependent histone gene transcription factors. J Cell Biochem. 1995;59:291–302. doi: 10.1002/jcb.240590302. [DOI] [PubMed] [Google Scholar]
- Stein GS, Lian JB, Dworetzky SI, Owen TA, Bortell R, Bidwell JP, van Wijnen AJ. Regulation of transcription-factor activity during growth and differentiation: involvement of the nuclear matrix in concentration and localization of promoter binding proteins. J Cell Biochem. 1991;47:300–305. doi: 10.1002/jcb.240470403. [DOI] [PubMed] [Google Scholar]
- Suske G. The Sp-family of transcription factors. Gene. 1999;238:291–300. doi: 10.1016/s0378-1119(99)00357-1. [DOI] [PubMed] [Google Scholar]
- van den Ent FM, van Wijnen AJ, Last TJ, Bortell R, Stein JL, Lian JB, Stein GS. Concerted control of multiple histone promoter factors during cell density inhibition of proliferation in osteosarcoma cells: reciprocal regulation of cell cycle-controlled and bone-related genes. Cancer Res. 1993;53:2399–2409. [PubMed] [Google Scholar]
- van der Meijden CMJ, Vaughan PS, Staal A, Albig W, Doenecke D, Stein JL, Stein GS, van Wijnen AJ. Selective expression of specific histone H4 genes reflects distinctions in transcription factor interactions with divergent H4 promoter elements. Biochim Biophys Acta. 1998;1442:82–100. doi: 10.1016/s0167-4781(98)00147-x. [DOI] [PubMed] [Google Scholar]
- van Wijnen AJ, Ramsey-Ewing AL, Bortell R, Owen TA, Lian JB, Stein JL, Stein GS. Transcriptional element H4-site II of cell cycle regulated human H4 histone genes is a multipartite protein/DNA interaction site for factors HiNF-D, HiNF-M, and HiNF-P: involvement of phosphorylation. J Cell Biochem. 1991;46:174–189. doi: 10.1002/jcb.240460211. [DOI] [PubMed] [Google Scholar]
- van Wijnen AJ, van den Ent FM, Lian JB, Stein JL, Stein GS. Overlapping and CpG methylation-sensitive protein-DNA interactions at the histone H4 transcriptional cell cycle domain: distinctions between two human H4 gene promoters. Mol Cell Biol. 1992;12:3273–3287. doi: 10.1128/mcb.12.7.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Ikura T, Eto K, Ota MS. Dynamic interaction of p220(NPAT) and CBP/p300 promotes S-phase entry. Biochem Biophys Res Commun. 2004;325:1509–1516. doi: 10.1016/j.bbrc.2004.10.198. [DOI] [PubMed] [Google Scholar]
- Wei Y, Jin J, Harper JW. The cyclin E/Cdk2 substrate and Cajal body component p220(NPAT) activates histone transcription through a novel LisH-like domain. Mol Cell Biol. 2003;23:3669–3680. doi: 10.1128/MCB.23.10.3669-3680.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie RL, Gupta S, Miele A, Shiffman D, Stein JL, Stein GS, van Wijnen AJ. The tumor suppressor interferon regulatory factor 1 interferes with SP1 activation to repress the human CDK2 promoter. J Biol Chem. 2003;278:26589–26596. doi: 10.1074/jbc.M301491200. [DOI] [PubMed] [Google Scholar]
- Ye X, Wei Y, Nalepa G, Harper JW. The cyclin E/Cdk2 substrate p220(NPAT) is required for S-phase entry, histone gene expression, and Cajal body maintenance in human somatic cells. Mol Cell Biol. 2003;23:8586–8600. doi: 10.1128/MCB.23.23.8586-8600.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Dynlacht B, Imai T, Hori T, Harlow E. Expression of NPAT, a novel substrate of cyclin E-CDK2, promotes S- phase entry. Genes Dev. 1998;12:456–461. doi: 10.1101/gad.12.4.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Kennedy BK, Lawrence BD, Barbie DA, Matera AG, Fletcher JA, Harlow E. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 2000;14:2283–2297. [PMC free article] [PubMed] [Google Scholar]