Abstract
Real-time immuno-PCR (RT-IPCR) is a powerful technique that combines ELISA with the specificity and sensitivity of PCR. RT-IPCR of phage-displayed peptides exploits the unique physical associations between phenotype (the displayed peptide) and genotype (the encoding DNA) within the same phage particle. Previously, we identified phage peptides specific for recombinant antibodies (rAbs) prepared from clonally expanded plasma cells in multiple sclerosis (MS) cerebrospinal fluid (CSF) and subacute sclerosing panencephalitis (SSPE) brain. Herein, we applied phage-mediated RT-IPCR to study reactivity of these specific phage peptides for the rAbs. Compared to standard ELISA, which required greater than 104 or 105 phage particles to detect binding to rAbs, RT-IPCR detected binding with as few as 100 phage particles. RT-IPCR was also superior to ELISA in determining relative affinities of rAbs for phage peptides and was effective in screening MS CSF for IgG reactivity to phage peptides. Phage-mediated RT-IPCR is a rapid, high-throughput technology that avoids the requirement for synthetic peptides and will facilitate the identification of candidate peptides that react with the IgG in MS CSF.
Keywords: Immuno-PCR, Phage peptide, Recombinant antibody
1. Introduction
In MS, there is increased IgG in brain and CSF, and the nature of the antigen against which the oligoclonal IgG is directed remains unknown. Our overall goal is to identify the antigen specificity of the oligoclonal IgG in patients with MS. We previously generated recombinant antibodies (rAb) from clonal populations of single plasma cells in the CSF of patients with multiple sclerosis (MS) (Owens et al., 2003, Ritchie et al., 2004) and in brain of a patient with subacute sclerosing panencephalitis (SSPE), chronic encephalitis caused by measles virus. These antibodies successfully identified epitopes/mimotopes from phage-displayed random peptide libraries (Yu et al., 2006 a, b; Owens et al., 2006). Such specific phage peptides have the potential to identify corresponding MS antigens. Initially, ELISA was used to determine phage peptide reactivity to the rAbs. Phage peptides with low to intermediate values were found, suggesting a need for improved techniques sensitive enough to identify low-affinity antibodies or low abundance of surrogate antigens.
Immuno-PCR (IPCR) uses PCR to detect specific proteins (Sano et al., 1992), relying on the affinity of DNA-labeled antibodies to bind specific antigens. During the exponential phase of PCR, the amount of product amplified reflects the amount of target antibodies bound by the antigens. Use of IPCR has successfully detected antigens associated with autoimmune diseases (Komatsu et al., 2001), and both pathogenic bacterial antigens (Wu et al., 2001; Liang et al., 2003) and bacterial toxins (Chao et al., 2004; Allen et al., 2006). Because phage particles exhibit the unique feature of a physical association between phenotype (the displayed peptide) and genotype (the encoding DNA) within the same particle, we applied phage to real-time (RT) IPCR. In this technique, the antibody-bound phage peptide is used as both detecting antigen and PCR template. Herein we report the application of phage-mediated RT-IPCR for detection of phage peptide binding to MS rAbs, for determination of relative affinities of rAb, and for rapid screening of MS CSF IgG reactive to phage peptides.
2. Materials and methods
2.1. CSF
CSF was obtained from patients with MS and other inflammatory central nervous system diseases with approval by the Institutional Review Board of the University of Colorado School of Medicine.
2.2. rAbs and phage
rAbs 02−19 #52 and 03−1 #37 cloned from the CSF of two MS patients, rAb2B4 cloned from an SSPE brain, and nine specific phage peptides panned by the rAbs were used in this study (Table 1). Features of these rAbs and phage peptides have been described (Burgoon et al., 1999; Yu et al., 2006 a, b; Owens et al., 2006).
Table 1.
rAbs and phage
| Source of rAbs | Panning rAb | Specific phage peptides |
|---|---|---|
| MS (MS02−19) CSF | #52 | 3−3−5 |
| MS (MS03−1) CSF | #37 | 2−6−1 |
| 2−6−4 | ||
| 2−6−12 | ||
| 2−6−17 | ||
| 3−6−8 | ||
| 3−6−6 | ||
| 3−612c | ||
| 3−6−18c | ||
| SSPE brain | 2B4 | 2B4-N |
| Random peptide library | Negative phage |
2.3. ELISA
Wells of ELISA plates were coated with rAb in 0.1 M carbonate buffer (100 μl, 1 μg/ml) overnight at 4°C, blocked with 3% BSA in TBS for 2 h at room temperature, and incubated for 1 h with various concentration of phage in TBS. After washing with 0.05% Tween 20-TBS, wells were incubated with mouse anti-M13 IgG-HRP antibody (New England BioLab, Beverly, MA) at 1:500 dilution for 1 h, and bound phage were detected with the peroxidase substrate ABTS (Zymed Laboratories Inc., San Francisco, CA). After incubation with the substrate for 20−30 min, absorbance at 415 nm was determined using a microplate reader (BioRad). All samples were tested in duplicate in at least 2 independent experiments. For phage RT-IPCR, phage bound in wells of ELISA plates were lysed and collected by adding 50 μl of deionized water to each well and heating at 95°C for 15 min. Phage in solution (4 μl) was used as template PCR amplification. For sandwich ELISA-PCR, wells of ELISA plates were coated overnight blocked with 3% BSA for 2 h, and incubated with 45 μl of MS and control CSF at a concentration of 5 μg IgG/ml for 2 h. Phage binding, washing and lysis were as described above.
2.4. Real-time PCR
Specific primers and probe (5′-FAM and 3′-TAMRA) for M13 phage real-time PCR have been described (Jaye et al., 2003). All real-time PCR was performed in an Applied Biosystems 7500 Fast Real-Time PCR system. Each PCR reaction (20 μl) consisted of 1X power SYBR® Green master mix, 750 nM of each primer and 4 μl of phage template. A dissociation-curve analysis was performed at 95°C for 15 sec, 60°C for 1 min and 95°C for 15 sec. Except when noted, all real-time PCR was performed using power SYBR® Green master mix. For PCR with TaqMan probe, primer concentration was 875 nM and probe concentration was 250 nM. A control reaction without template was included in each run. The thermal cycle conditions included 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 45 sec. The 100-bp amplicon encoding the α-complementing fragment of the Lac Z gene of the phage was confirmed by electrophoresis.
3. Results
We previously identified phage peptides that were specific for MS rAbs prepared from clonally expanded plasma cells in MS CSF (Yu et al., 2006 a, b). Here we combined real-time PCR with standard ELISA to characterize the interaction of these phage with MS rAbs, and to use these phage to screen MS CSF for IgG reactivity.
3.1. Amplification of M13 phage DNA using real-time PCR
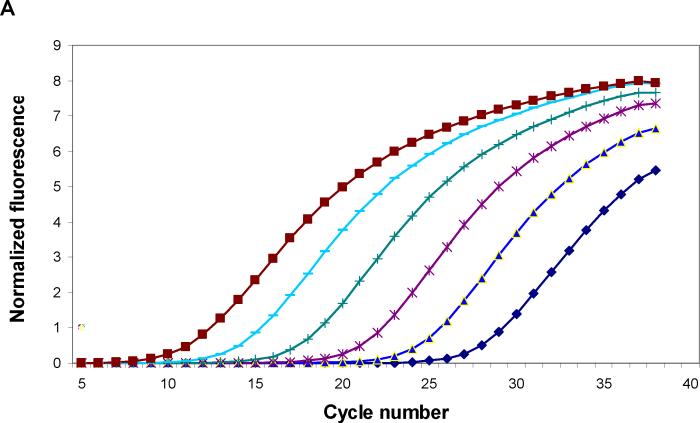
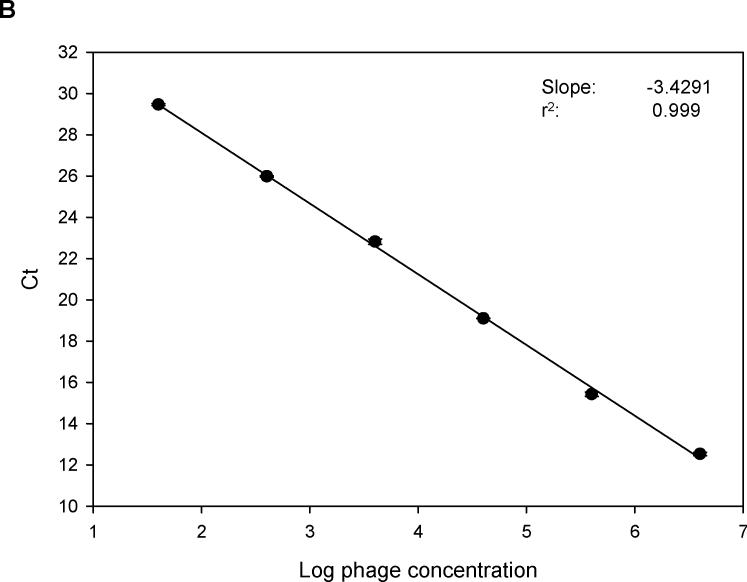
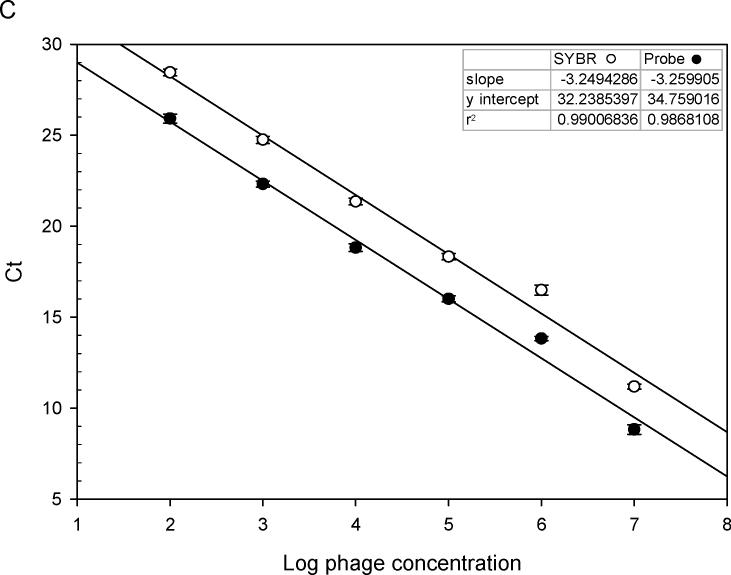
Specific primers and probe of M13 phage were synthesized as described (Jaye et al., 2003). Use of the 7500 Fast Real-time PCR system enabled amplification of phage DNA with whole phage particles rather than purified single-strand phage DNA. With power SYBR® Green master mix and phage primers, 10-fold serial dilutions of phage DNA were amplified. Fig. 1A shows a typical amplification plot during 40 cycles. When cycle threshold values were plotted against log10 phage number, a linear regression curve was generated across a wide range of phage dilutions (r2=0.999. Fig.1B). Compared to amplifications with phage TaqMan probe, PCR amplification of phage DNA using SYBR Green dye was more sensitive (Fig. 1C).
Fig. 1.
(A). Real-time PCR amplification of 10-fold serial dilutions of a M13 phage. Primers specific for α-complementing sequences in the LacZ gene of M13 phage were used with power SYBR® Green master mix for PCR amplification. The intensity of SYBR green fluorescence was plotted against the cycle number. Each curve shows the amplification profile for input phage from 1 × 107 (far left) to 1 × 102 pfu (far right) per PCR reaction. (B) Standard curve of real-time PCR amplification of M13 phage (peptide 3−3−5) with power SYBR® Green master mix. Cycle threshold (Ct) plotted against the log10 phage concentration reveals a linear correlation. Regression parameters are shown on top right. Each data point is the average of two independent measurements. (C) Comparison of standard curves of M13 phage real-time PCR with SYBR Green master mix and with an M13 phage-specific TaqMan probe (5′-FAM and 3′-TAMRA). As shown on the y-intercept (SYBR: 32.23; TaqMan: 34.76), PCR with SYBR is more sensitive than that with TaqMan probe. Linear regression parameters are shown on top right. Each data point is the average of two independent measurements.
3.2. Phage RT-IPCR increases detection sensitivity
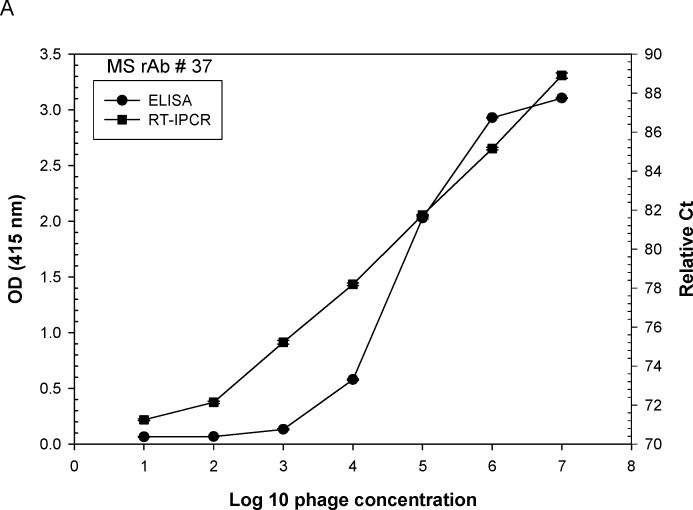
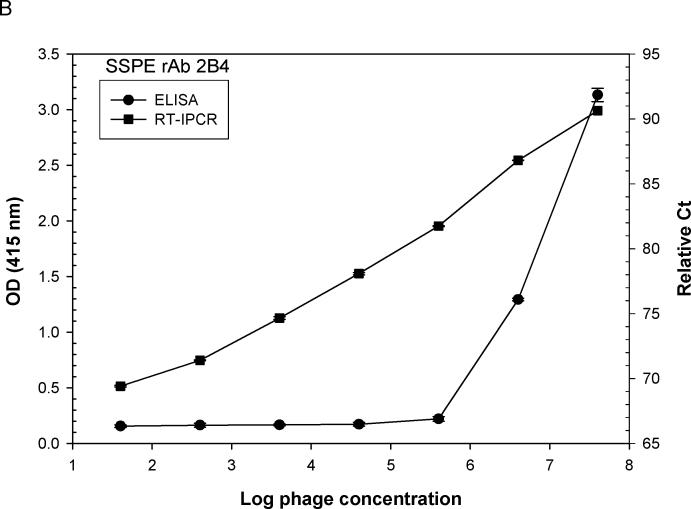
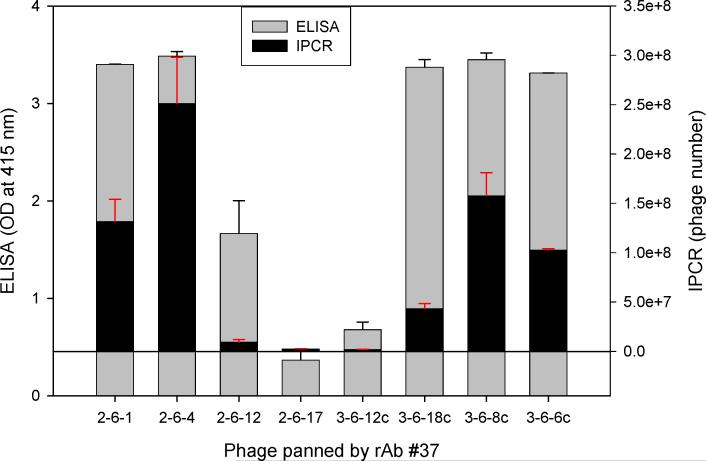
Ten-fold serial dilutions of phage peptides were added to ELISA plates pre-incubated with rAbs overnight at 4°C. For phage RT-IPCR, bound phage were lysed and collected by heating the ELISA plate at 95°C for 15 min to release phage DNA, and 4 μl of the phage lysate was used directly as template for PCR amplification. For standard ELISA, bound phage were incubated with mouse anti-M13 IgG-HRP antibody and detected with the peroxidase substrate ABTS. Using MS rAb #37 and SSPE rAb 2B4, each with their corresponding phage peptides, as little as 100 eluted phage particles were detected. Phage RT-IPCR detected phage particles in the range of 102 to 107, whereas standard phage ELISA detected phage only when 104 to 105 or greater phage were added to the ELISA wells (Fig. 2A, B). Further analysis of eight phage previously shown to be specific for MS rAb #37 (Yu et al., 2006 a, b) revealed a wide range of binding affinities to the rAb. The OD values of phage 2−6−1, 2−6−4, 3−6−12c, 3−6−8c and 3−6−6c were similar in ELISA, but the number of bound phage revealed with IPCR showed distinct differences (Fig. 3).
Fig. 2.
Comparison of sensitivity of phage binding to recombinant antibody (rAb) by (RT) IPCR vs. standard ELISA. ELISA plates coated with 50 ng of either MS rAb #37 (A) or SSPE rAb 2B4 (B) were incubated with 10-fold serial dilutions of phage (phage 2−6−4 is specific for rAb #37; phage 2B4-N is specific for rAb 2B4). Phage binding was assayed using either (RT) PCR or standard ELISA with HRP-conjugated anti-M13 antibody as described in Materials and methods. The ELISA OD values and PCR relative Ct values (100 minus Ct) were plotted against input phage concentrations. Each data point is the average of two independent measurements. RT-IPCR detected as little as 100 bound phage particles compared to standard ELISA which required greater than 104 phage particles for signal detection with rAb #37 (A) or 105 phage particles with rAb 2B4 (B).
Fig. 3.
Comparison of (RT) IPCR and traditional ELISA in determining phage binding to a single MS rAb. Eight different phage (5 × 108) were added to ELISA wells pre-coated with 50 ng of MS rAb #37 and binding was assayed by RT-IPCR or standard ELISA using anti-M13 antibody as described in the Fig. 2 legend. White and black bars show ELISA OD values and IPCR bound phage number, respectively, for the same phage population. With 5 × 108 phage, ELISA values for phage 2−6−1, 2−6−4, 3−6−18c, 3−6−8c and 3−6−6c were similar, in contrast to distinct differences in the number of bound phage revealed by IPCR. Error bars represent standard deviation of duplicate samples.
3.3. Binding affinity of phage to rAbs
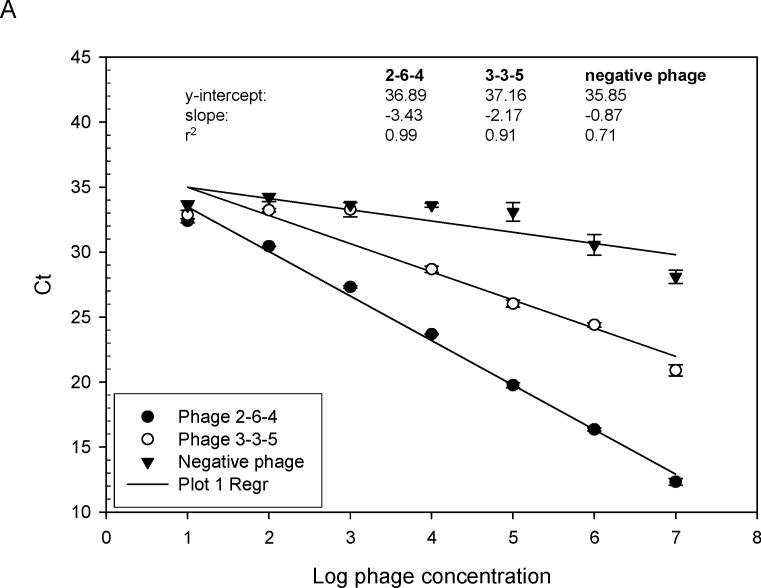
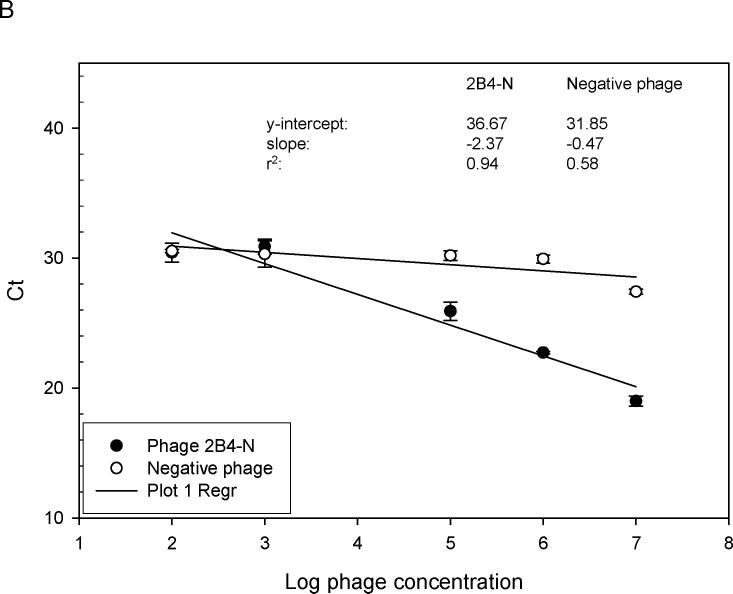
Previous screening of phage-displayed random peptide libraries with MS rAbs identified phage peptides that bound to the rAbs with varying ELISA values (Yu et al., 2006 b). To quickly and accurately determine the relative affinity of selected phage peptides, quantitation by RT-IPCR was used to compare bindings of different phage peptides. As shown in Fig. 4A, MS rAb #37, previously found to have high affinity for phage peptide 2−6−4 (Yu et al., 2006 a, b), had a lower slope value than that of MS rAb #52, previously shown to have lower affinity for phage peptide 3−3 5 (Yu et al., 2006 a, b). With as few as 100 phage particles, binding to rAb could be determined. The described high-affinity binding of SSPE rAb 2B4 to phage 2B4-N (Owens et al., 2006) was confirmed (Fig. 4B).
Fig. 4.
Relative binding affinity of rAbs to phage using RT-IPCR. (A). MS rAbs #37 and #52 were coated onto wells of ELISA plates overnight at 4°C. Ten-fold serial dilutions of phage 2−6−4 (previously shown to be specific for rAb #37) and an irrelevant phage were added to wells containing rAb #37; 10-fold serial dilutions of phage 3−3−5 (previously shown to be specific for rAb #52) were added to wells containing rAb #52. Phage binding was determined by (RT) PCR. High-affinity rAb #37 had lower Ct values (slope = −3.43) than the lower affinity rAb #52 (slope = −2.17), and the negative control phage had the highest Ct values (slope = −0.87). B. SSPE rAb 2B4 binding to phage 2B4-N (previously shown to be specific for 2B4-N). High-affinity 2B4 had a lower Ct value (slope = −2.37) compared to the negative control phage with high Ct value (slope = −0.47). Error bars represent standard deviation of duplicate samples.
3.4. Phage RT-IPCR screening of MS CSF for reactive IgG
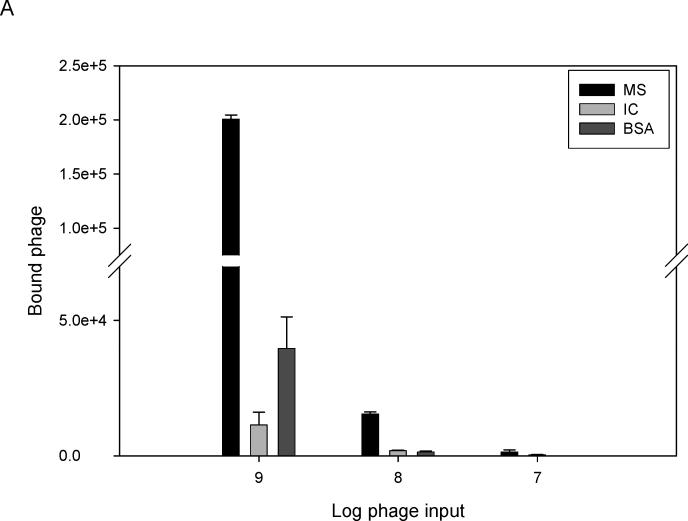
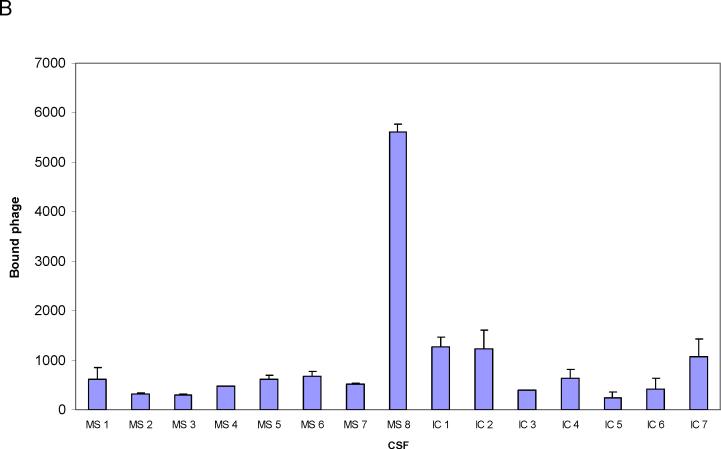
A sandwich ELISA was prepared using anti-human IgG-coated ELISA plates incubated with CSF from one MS patient (IgG concentration, 5 μg/ml). Phage 2−6−4 (previously identified by panning with MS rAb #37) at three different concentrations were added to bound MS IgG, and the number of bound phage were determined by (RT) PCR. High concentrations of phage (109 and 108) were required to show binding of phage peptides to MS IgG (Fig. 5A). Phage 2−6−4 (108 per well) was tested with eight MS CSFs; only MS 03−1 CSF from which the panning rAb #37 was cloned contained IgG that reacted with this phage peptide 2−6−4 (Fig. 5B). Note that phage RT-IPCR of CSF identified specific IgG in CSF without synthesis or purification of peptides.
Fig. 5.
(A) Detection of phage peptide-specific IgG in MS CSF by RT-IPCR. MS CSF (03−1), CSF from an inflammatory control patient (IC, Cryptococcal meningitis), both at IgG concentrations of 5 μg/ml, and BSA were added to ELISA plates coated with anti-human IgG. Phage 2−6−4 (specific for rAb #37) at concentrations of 109, 108 and 107 were added to each well. Bound phage was collected and quantified by (RT) PCR as described in Fig. 2A. Phage peptide 2−6−4 bound specifically to IgG in MS CSF, but not to IC control CSF or to BSA. Error bars represent standard deviation of duplicate samples. (B) MS CSF screening for phage-specific IgG using RT-IPCR. Eight MS CSFs and 7 inflammatory control CSFs were screened with phage 2−6−4 (5 × 108) for IgG reactivity as described in Fig 5A. Phage 2−6−4 bound only to MS 03−1 CSF (MS 8) from which rAb #37 was cloned, but not to any other MS CSFs or the IC CSF. Error bars represent standard deviation of duplicate samples.
4. Discussion
IgG is increased in brain and CSF of MS patients, and is expressed in a restricted oligoclonal pattern. The nature of the antigen against which the oligoclonal IgG is directed remains unknown. Studies to determine the nature of the specific antigen have exploited the fact that clonal populations of plasma cells and B cells are present in the CSF of patients with MS (Qin et al 1998; Owens et al., 2003, Ritchie et al., 2004). Monoclonal rAbs prepared from overrepresented immunoglobulin sequences found in these cells have been used to search for antigen specificity. Screening phage-displayed random peptide libraries with rAbs has been one of the approaches used to identify antigen specificity.
Previous use of ELISA to determine phage peptide reactivity to MS rAbs (Yu et al., 2006 a, b) identified phage peptides with low to intermediate values, suggesting a need for improved techniques sufficiently sensitive to detect low-abundance antigens or low-affinity antibodies. Here, we find that compared to standard ELISA which required greater than 104 or 105 phage particles to detect binding to rAbs, RT-IPCR detected binding with as little as 100 phage particles and more than 5 log units of phage were detected with high reproducibility.
RT-IPCR was also superior to ELISA in determining relative affinities of rAbs for phage peptides. Studies of antibody affinity have been useful in distinguishing present from previous exposure to disease-causing agents (Giovannoni et al., 2006). Use of RT-IPCR identified rAbs with differing affinities to phage peptides based on the linear correlation between phage number and cycle threshold (Fig. 4A). Furthermore, as few as 100 phage particles could be used to determine the relative affinity of a rAb coated on wells of an ELISA plate.
Importantly, RT-IPCR was successful in screening MS CSF from multiple patients for IgG reactivity to phage peptides. The sandwich ELISA-PCR we used avoids the need for synthesis of each candidate peptide and instead uses phage directly in the assay, saving time and resources while increasing detection efficiency. Because peptide epitopes/mimotopes can be identified by various target molecules (protein, lipid and carbohydrate), phage (RT) IPCR provides a universal detecting method for determining reactivity of peptides regardless of the nature of the targets. These peptides can be used to screen bioinformatics databases to recognize cognate antigens, survey CSFs from patient populations, identify shared immune response, and measure the relative binding strength of IgG in disease states.
Overall, compared to standard ELISA, phage RT-IPCR is a highly sensitive and more efficient technique to rapidly characterize the interaction between phage peptides and antibody. Application of RT-IPCR to phage peptides identified by rAbs prepared from clonal populations of antibody-producing cells in chronic inflammatory diseases of unknown etiology has the potential to identify disease-relevant antigens.
Acknowledgements
We thank Dr. Greg Owens for providing rAbs and critical review of this manuscript, Marina Hoffman for editorial review and Cathy Allen for preparing the manuscript. This work was supported by grant NS 32623 from the National Institutes of Health.
Abbreviations:
- RT-IPCR
real-time immuno-PCR
- rAb
recombinant antibody
- MS
multiple sclerosis
- SSPE
subacute sclerosing panencephalitis
- CSF
cerebrospinal fluid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allen RC, Rogelj S, Cordova SE, Kieft TL. An immuno-PCR method for detecting Bacillus thuringiensis Cry1Ac toxin. J. Immunol. Methods. 2006;308:109. doi: 10.1016/j.jim.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Burgoon MP, Williamson RA, Owens GP, Ghausi O, Bastidas RB, Burton DR, Gilden DH. Cloning the antibody response in humans with inflammatory CNS disease: isolation of measles virus-specific antibodies from phage display libraries of a subacute sclerosing panencephalitis brain. J. Neuroimmunol. 1999;94:204. doi: 10.1016/s0165-5728(98)00243-4. [DOI] [PubMed] [Google Scholar]
- Chao HY, Wang YC, Tang SS, Liu HW. A highly sensitive immunopolymerase chain reaction assay for Clostridium botulinum neurotoxin type A. Toxicon. 2004;43:27. doi: 10.1016/j.toxicon.2003.10.013. [DOI] [PubMed] [Google Scholar]
- Giovannoni G, Chapman MD, Thompson EJ. The role of antibody affinity for specific antigens in the differential diagnosis of inflammatory nervous system disorders. J. Neuroimmunol. 2006;180:29. doi: 10.1016/j.jneuroim.2006.06.033. [DOI] [PubMed] [Google Scholar]
- Jaye DL, Nolte FS, Mazzucchelli L, Geigerman C, Akyildiz A, Parkos CA. Use of real-time polymerase chain reaction to identify cell- and tissue-type-selective peptides by phage display. Am. J. Pathol. 2003;162:1419. doi: 10.1016/S0002-9440(10)64275-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Kobayashi D, Saito K, Furuya D, Yagihashi A, Araake H, Tsuji N, Sakamaki S, Niitsu Y, Watanabe N. Tumor necrosis factor-alpha in serum of patients with inflammatory bowel disease as measured by a highly sensitive immuno-PCR. Clin. Chem. 2001;47:1297. [PubMed] [Google Scholar]
- Liang H, Cordova SE, Kieft TL, Rogelj S. A highly sensitive immuno-PCR assay for detecting Group A Streptococcus. J. Immunol. Methods. 2003;279:101. doi: 10.1016/s0022-1759(03)00247-3. [DOI] [PubMed] [Google Scholar]
- Owens GP, Ritchie AM, Burgoon MP, Williamson RA, Corboy JR, Gilden DH. Single-cell repertoire analysis demonstrates that clonal expansion is a prominent feature of the B cell response in multiple sclerosis cerebrospinal fluid. J. Immunol. 2003;171:2725. doi: 10.4049/jimmunol.171.5.2725. [DOI] [PubMed] [Google Scholar]
- Owens GP, Shearer AJ, Yu X, Ritchie AM, Keays KM, Bennett JL, Gilden DH, Burgoon MP. Screening random peptide libraries with subacute sclerosing panencephalitis brain-derived recombinant antibodies identifies multiple epitopes in the C-terminal region of the measles virus nucleocapsid protein. J. Virol. 2006;80:12121. doi: 10.1128/JVI.01704-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Duquette P, Zhang Y, Talbot P, Poole R, Antel J. Clonal expansion and somatic hypermutation of V(H) genes of B cells from cerebrospinal fluid in multiple sclerosis. J. Clin. Invest. 1998;102:1045. doi: 10.1172/JCI3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie AM, Gilden DH, Williamson RA, Burgoon MP, Yu X, Helm K, Corboy JR, Owens GP. Comparative analysis of the CD19+ and CD138+ cell antibody repertoires in the cerebrospinal fluid of patients with multiple sclerosis. J. Immunol. 2004;173:649. doi: 10.4049/jimmunol.173.1.649. [DOI] [PubMed] [Google Scholar]
- Sano T, Smith CL, Cantor CR. Immuno-PCR: very sensitive antigen detection by means of specific antibody-DNA conjugates. Science. 1992;258:120. doi: 10.1126/science.1439758. [DOI] [PubMed] [Google Scholar]
- Wu HC, Huang YL, Lai SC, Huang YY, Shaio MF. Detection of Clostridium botulinum neurotoxin type A using immuno-PCR. Lett. Appl. Microbiol. 2001;32:321. doi: 10.1046/j.1472-765x.2001.00909.x. [DOI] [PubMed] [Google Scholar]
- Yu X, Gilden DH, Ritchie AM, Burgoon MP, Keays KM, Owens GP. Specificity of recombinant antibodies generated from multiple sclerosis cerebrospinal fluid probed with a random peptide library. J. Neuroimmunol. 2006 a;172:121. doi: 10.1016/j.jneuroim.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Yu X, Owens GP, Gilden DH. Rapid and efficient identification of epitopes/mimotopes from random peptide libraries. J. Immunol. Methods. 2006 b;316:67. doi: 10.1016/j.jim.2006.08.006. [DOI] [PubMed] [Google Scholar]